我于2007年回国,下定决心做与自己之前在美国不同、也尽可能与众不同的研究。
当时除了表面发几篇文章,大量时间精力用于探索和铺开了几条长线研究。
同时并举经典和现代遗传学,辅以人脑核磁共振成像,研究人类高级认知,始于2007年回国之前一次访问北京师范大学,此后布局,用了六千多个被试者,其中几个家系中有一个几十人的大家系(含18位脸盲者)。文章到2024年3月18日被Genetics杂志接受,3月28日线上发表。
人脸识别,是高级社会认知、也是高级视觉认知。
脸识别的人类遗传学:脸盲人中发现MCTP2基因突变
孙芸, 门卫伟, Ingo Kennerknecht, 方万, 郑厚峰, 张文霞,饶毅
首都医科大学 首都医学科学创新中心,北京脑科学研究所,北大清华生命科学研究中心、北大IDG/McGovern脑研究所、北大生命科学学院,深圳湾实验室分子生理学研究所,北大交叉学院核磁共振成像中心、北京医学物理与工程重点实验室,北大物理学院重离子研究所,德国Westfälische Wilhelms大学人类遗传研究所,西湖大学生命科学学院
摘要
脸识别对于视觉认知和社会认知都很重要。虽然脸盲(prosopagnosia或face blindness)已经被发现了七十多年,脸特异反应神经元已被发现近半个世纪,其分子遗传学机理不清楚。在此我们报道用经典遗传学和现代基因组学研究17年的结果。从一个含18位在日常生活脸识别困难的先天脸盲(congenital prosopagnosia, CP)成员的家系,我们在MCTP2基因找到完全共分离的独家突变,它编码脑内表达的一个钙结合的跨膜蛋白。在更多CP家族、以及6589人中筛查到的CP中发现更多MCTP2 的罕见突变。14位MCTP2基因移码突变S80fs 的携带者,与19位非携带者在脸识别有显著差别。包括一个家庭有10位成员的6个家庭,显示S80fs与CP的相关性。功能核磁共振成像发现MCTP2突变的CP对个体脸识别的障碍相关于脑内右侧纺锤体脸识别区域对重复脸的重复压制减低。最早发现脸盲的76年后、第一个脸盲病人发现47年后,我们的结果揭示了MCTP2 突变致CP的基因罹患性。这是第一次发现人类高级视觉社会认知所必需的基因。
引言
虽然人类认知非常有趣,对人类认知的分子研究却罕见。通常,先在动物研究某些基因,然后在人类研究它们。这种途径限制了研究在动物必需有的表型,而且常常是低等动物,因为在非人灵长类动物很难通过功能研究发现基因。因此,对于只在人类存在的认知、或在低等动物不存在的认知进行分子研究,远远落后于对于简单行为的分子研究。
但是,对人类疾病的遗传研究非常成功(Gusella et al., 1983; Schrott et al., 1972; Tsui et al., 1985)。 对于用遗传学研究的可行性来说,没有必要假定人类认知与疾病有根本的差别。遗传学提供了可以研究人类认知的无创途径。近十年来,我们实验室进行了几次人类认知的全基因组相关研究(GWAS),从记忆、社会从众性、到视觉认知的感知切换和下行控制(Chen et al., 2018a; Chen et al., 2018b; Zhu et al., 2019; Zhu et al., 2016; Zhu et al., 2018; Zhu et al., 2020)。虽然我们发现了相关的标记,但我们不知道涵盖标记的基因、或标记附近的基因与所研究的认知有没有因果关系。遗传学上,大家系的连锁分析成功地找到人类疾病的基因突变。我们用此途径研究脸识别的分子遗传基础,研究结果在此。
脸识别,视觉识别最复杂之一,为社会认知所必需,通常但不总是在高等动物(Bruce and Young, 1986; Freiwald et al., 2016; Grill-Spector and Malach, 2004; Tsao and Livingstone, 2008)。 半个世纪以来,在猴的下颞叶皮层(IT)(Desimone et al., 1984; Gross et al., 1972; Rolls, 1984; Yamane et al., 1988)和颞上沟(STS)(Baylis et al., 1985; Bruce et al., 1981; Chitty et al., 1985; Perrett et al., 1987; Perrett et al., 1982; Perrett et al., 1984, 1985; Rolls, 1984; Rolls et al., 1985; Saito et al., 1986)发现了对脸有特异反应的神经元。电刺激猴脑的实验结果支持了神经元在脸识别的功能意义(Afraz et al., 2006; Moeller et al., 2017)。神经元的脸特异反应也在人的事件相关电位中(Allison et al., 1999; McCarthy et al., 1999; Puce et al., 1999)或直接电记录(Kreiman et al., 2000)检测到。枕叶和颞叶对脸特异反应的脑区也通过正电子扫描(PET)(Haxby et al., 1994; Sergent et al., 1992)和概念和磁共振成像(fMRI)(Barraclough and Perrett, 2011; Clark et al., 1996; Dzhelyova et al., 2011; Kanwisher et al., 1997; Liu et al., 2010; Malach et al., 1995; McCarthy et al., 1997; Puce et al., 1996; Tsao et al., 2003; Tsao et al., 2006; Tsao et al., 2008)在人脑中检测到。跨颅磁刺激的结果也与这些区域对脸识别功能重要的可能性一致(Pitcher et al., 2009; Pitcher et al., 2008; Polk et al., 2007)。
脸盲(prosopagnosia), 首先为德国神经内科医生Joachim Bodamer所报道(Bodamer, 1947), 是脸识别的缺陷,不是由于低层视觉或高层语义问题。迄今为止,在分子水平,对脸盲或脸识别都一无所知。
先天脸盲(congenital prosopagnosia, CP), 也称为发育性或遗传性脸盲(OMIM 610382), 最初在1976年被报道(McConachie, 1976)是对脸的视觉学习和识别的选择性损害,缺乏任何可以检测到的神经损伤(Behrmann and Avidan, 2005; Damasio et al., 1990; Duchaine and Nakayama, 2006b; Gruter et al., 2008; Kress and Daum, 2003; McConachie, 1976; Nunn et al., 2001; Susilo and Duchaine, 2013)。问卷式的筛选方法(Kennerknecht, 2021; Kennerknecht et al., 2006; Kennerknecht et al., 2008a; Kennerknecht et al., 2007)与行为测试(Bowles et al., 2009)都估计一般人群CP发病率在1.8 to 2.9%, 全球估计数千万CP个体。家系研究(De Haan, 1999; Dobel et al., 2007; Duchaine et al., 2007; Galaburda and Duchaine, 2003; Grueter et al., 2007; Johnen et al., 2014; Kennerknecht et al., 2006; Kennerknecht et al., 2008b; Kennerknecht et al., 2007; Lee et al., 2010; McConachie, 1976; Schmalzl et al., 2008)和双生子研究(McKone and Palermo, 2010; Polk et al., 2007; Wilmer et al., 2010; Zhu et al., 2010)提示CP与脸识别能力具有高度遗传性。家谱分析观察到简单的常染色体显性遗传模式(De Haan, 1999; Duchaine et al., 2007; Grueter et al., 2007; Kennerknecht et al., 2006; Lee et al., 2010; Schmalzl et al., 2008), 显示单个基因的突变可以导致脸识别缺陷。
我们从一个含18位日常生活中识别脸困难的大家系出发,发现了一个CP 罹患基因,编码“多重 C2 区域跨膜2”蛋白质(MCTP2, GenBank:NM_018349)。在日常生活脸识别困难的人中找到更多的MCTP2 罕见突变和相关性。fMRI结果显示MCTP2突变CP患者对个体脸识别缺陷 相关于右侧纺锤体脸区域(rFFA)对同样脸的重复呈现反应异常。我们的发现可以刺激用遗传学和基因组学研究人类高级认知。
结果
三代含CP患者的家系
面试了一个三代家庭(家庭A)的35位成员(表1, 附表S1, 图1a)。18家庭成员明显在日常生活中有脸识别困难:9位(IV:2, IV:4, V:9, V:10, V:11, V15, VI:5, VI:6 and VI:9)在我们联系之前就知道他们早期生活就有这一问题;4位(IV:6, V:4, V:6 and VI:8)很长时间认人有可能但不知道原因; 5 位(IV:10, V:1, V:13, V:19 and VI:7)无视其日常生活中这一问题,而采取措施依赖脸之外线索的策略(表1)。自动注视接触需求与脸识别困难或用代偿策略之间有显著的负相关(p < 0.001, r = -0.658, n = 35)。
与以前研究(Johnen et al., 2014; Schmalzl et al., 2008)一致, 家庭内就有异质性,无论是剑桥脸记忆检测-中文(Cambridge Face Memory Test-Chinese, CFMT-C)(Bowles et al., 2009; McKone et al., 2012), 配对剑桥车记忆检测(the matched Cambridge Car Memory Test,CCMT)(Dennett et al., 2012)还是脸倒置效应(the Face Inversion Effect,FIE)区别检测(Yovel and Kanwisher, 2005)。14位在真实世界有脸识别困难的家庭成员中10位在至少一种行为测试中有缺陷。V:1, V:9, V:11, V:13, V:15, V:19和VI:8在CFMT-C得分中有一个分数低于正常水平的两个标准差(SDs), 而V:4(z = -1.60)和V:6(z = -1.61)行为也明显差(低于-1.5 SDs)。在检测整体加工的FIE区分试验, V:6(z = -1.62), V:9(z = -1.62), V:11(z = -2.56)和VI:5(z = -1.81)的导致效应缺陷。V:1(z = -2.96), V:13(z = -1.73), V:15(z = -1.55)和V:19(z = -1.52)区别正置脸的能力也很差。有四位(V:10, VI:6, VI:7和VI:9)日常有困难,但在脸识别的检测中表现无异。他们都在CCMT检测很好,显示他们识别其他物理刺激的能力正常,与其识别脸无关。每位成员的Z分见表1。
染色体15q上特异区域含候选CP罹患基因
为了错在过分小心,家庭A的初步连锁分析(称为连锁1,见方法)(附表S1),我们诊断9个家庭成员为CP患者(V:1, V:4, V:6, V:9, V:11, V:13, V:15, V:19 and VI:8),诊断标准不仅是面谈显示脸孔识别异常,而且还在CFMT行为检测表现差。最高LOD(logarithm of odds)分数为3.49, 提示可能有一个CP连锁基因,一个3.9兆碱基对(Mb)的主要候选区域(major candidate region,MCR)从rs12148885(chr15:94251330, 15q26.1)跨到rs288435(chr15:98160439, 15q26.2),对应于15号染色体上1-LOD 下降区, 依据于hg19组装(图1b)。基因组筛选结果显示于附图S3a。一千次重复的计算机模拟显示经验p为0.023, 提示这一结果在基因组水平有显著性。另一区域9q21.13 的LOD分数超过1.0(附图S3a), 但计算机模拟没有达到基因组显著性。MCR内同享的单倍型见附图S4a。
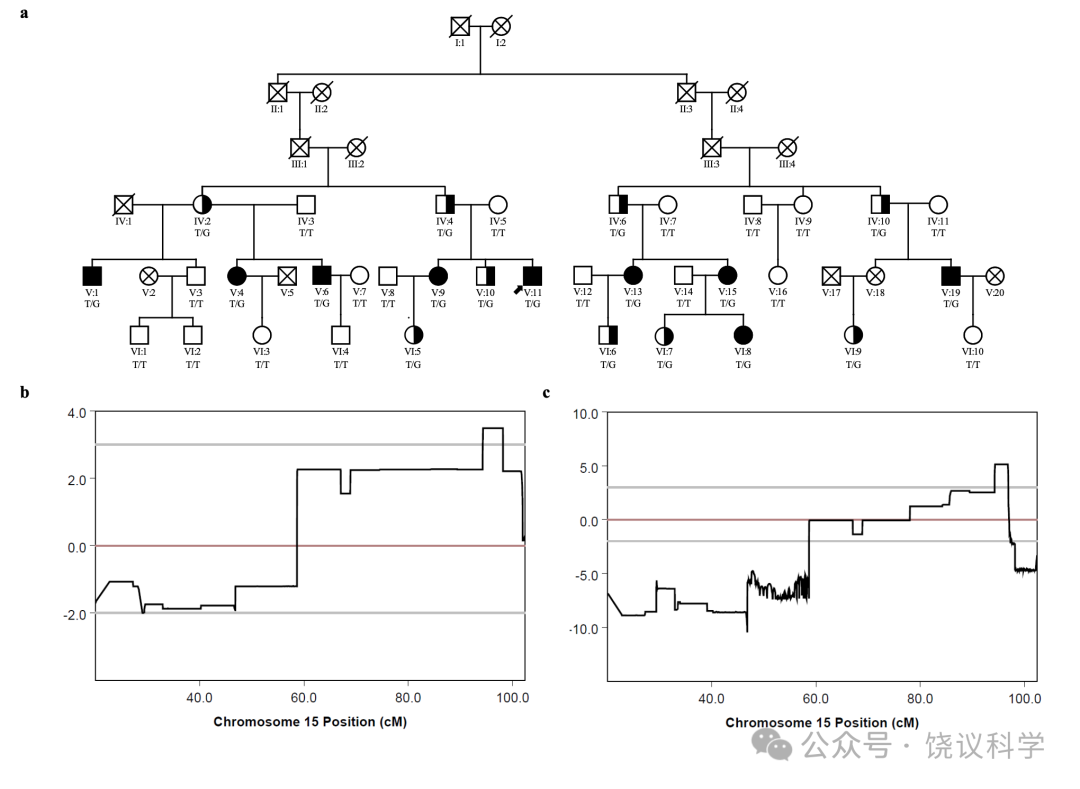
有可能有些患者用应对策略完成了个体识别、而不是脸识别,导致分类的模糊(Dalrymple and Palermo, 2016; Duchaine and Weidenfeld, 2003; Grueter et al., 2007)。因此,我们进行了第二次连锁分析(连锁2,见方法)。较连锁1在患者中增加了4位日常有脸识别问题而在行为检测表现正常的(V:10, VI:5, VI:6 and VI:7)和4位日常脸识别问题的先证者(IV:2, IV:4, IV:6 and IV:10)(表S1)。连锁2分析延伸了连锁1,继续支持rs6497114(chr15:94245722)和rs11045(chr15:96883321)之间的区域LOD分数高于3(最高LOD分数= 5.13), 跨染色体15q26.1至26.2之间的2.64 Mb(图1c, 附图S3b)。另一区域10q24.2的LOD 分数高于1。模拟后,15号染色体上最高LOD分数有统计显著性(p< 0.008)。MCR内同享的单倍型见附图S4b。
MCR内用最新NCBI资料库(https://www.ncbi.nlm.nih.gov/genome/gdv/)注释的候选基因列在附表S7。
全基因组测序揭示MCTP2 基因的一个突变
为了找到A家庭连锁CP的染色体15q26.1-26.2区域的影响脸识别的因果关系突变,我们对连锁1的9位受影响个体(V:1, V:4, V:6, V:9, V:11, V:13, V:15, V:19和VI:8, 表S1)做了全基因组测序(WGS)。MCR内, 染色体chr15:94983466上MCTP2只有一个变异(NM_018349.4:c.2147T>G, NP_060819.3:(p.I716S))在全部9位CP都是杂合体。这个异义突变(c.2147T>G)为A家庭特有,不存在于dbSNP(v150), ExAC03和genomAD(v2.1.1)几个资料库,虽然这一位置还报道过另外多等位单碱基多样性(SNP)位点rs200314451 :在东亚gnomAD v2.1.1库,NM_018349.4:c.2147T>C, NP_060819.3:p.I716T以最小等位基因频率(MAF)0.000存在,NM_018349.4:c.2147T>A, NP_060819.3:p.I716N以MAF 0.0001088存在。在我们扩大的3600 中国人队列中,我们没有检测到同一突变。
为了研究这一变异是否罹患CP的强候选者,我们在家系直接家系Sanger测序,并进行共分离分析。除了连锁分析1的9位, MCTP2的 c.2147T>G [p.I716S]也存在于4位脸识别能力弱、但因为年龄不合适用行为检测的前辈,也存在于CFMT-C检测正常但日常生活中识别脸能力弱的5位家庭成员(图1a, 半实符号)。
所以, MCTP2 编码NP_060819.3:p.I716S的突变看来是这一大CP家系所有有日常脸识别问题的病人在MCR唯一共享的功能变异。SIFT(Ng and Henikoff, 2001)预计I716S致病。MCTP2基因全长占基因组DNA的180千碱基对, 有22个编码的外显子, 编码的蛋白质有878 个氨基酸残基,分成三个C2区域和两个跨膜区(TMRs)(图S5a)。第一个TMR的NP_060819.3:I716在发展出很好脸加工的灵长类高度保守(附图S5a与S5b)(Freiwald et al., 2016; Hung et al., 2015; Moeller et al., 2008; Tsao et al., 2003; Tsao et al., 2006; Tsao et al., 2008)。
扩大筛选找到更多CP和CP家系含更多MCTP2 突变
在理论和实验考虑的基础上,罕见的功能等位基因被认为是表型遗传的主要贡献者(Cirulli and Goldstein, 2010; Heinzen et al., 2015; Jordan et al., 2010; Pritchard, 2001)。从典型的A家庭获得的结果显示罕见、甚至特有的MCTP2基因功能突变可能与CP表型有关。
在问卷筛选的第一个队列的2904人(见方法)中, 我们找到75位分数与平均至少差2 SDs并测了其MCTP2 外显子的序列。7位中发现了5个罕见杂合子功能变异体,包括一个移码突变和4给异义突变(图2a和附图S5a, 附表S8)。
MCTP2基因外显子1的移码突变(NM_018349.4:c.239delG, NP_060819.3:p.S80fs)见于报告脸识别困难的75位中的3位(个体B0001, B0009和B0046)(图2b)。进一步面试揭示所有3位携带p.S80fs的报告长期且恼人的脸识别主观不确定性,在其早年意识到这一问题(附表S2。B0001的一位亲戚携带p.S80fs,采取了观察其他特征的策略,这样她需要更长时间识别人(图2c, 附表S2)。
在个体B0011的家庭, 含外显子5的NP_060819.3:p.M272L突变的那些人报告他们自己认人的策略,但还是难于应对其困难(图2d与2e, 表S2)。外显子8的NP_060819.3:p.T374A突变见于B0010个体,自己认识到有严重的脸识别缺陷(图2f, 附表S2)。 B0003的家庭, 外显子12的NP_060819.3:p.V548I突变与表型相关性很好(图2g与2h, 附表S2)。B0002的家庭, NP_060819.3:p.R641Q突变与家庭所有有表型儿童共分离(图2i与2j)。这些突变位点在动物进化中保守(图S5b)。
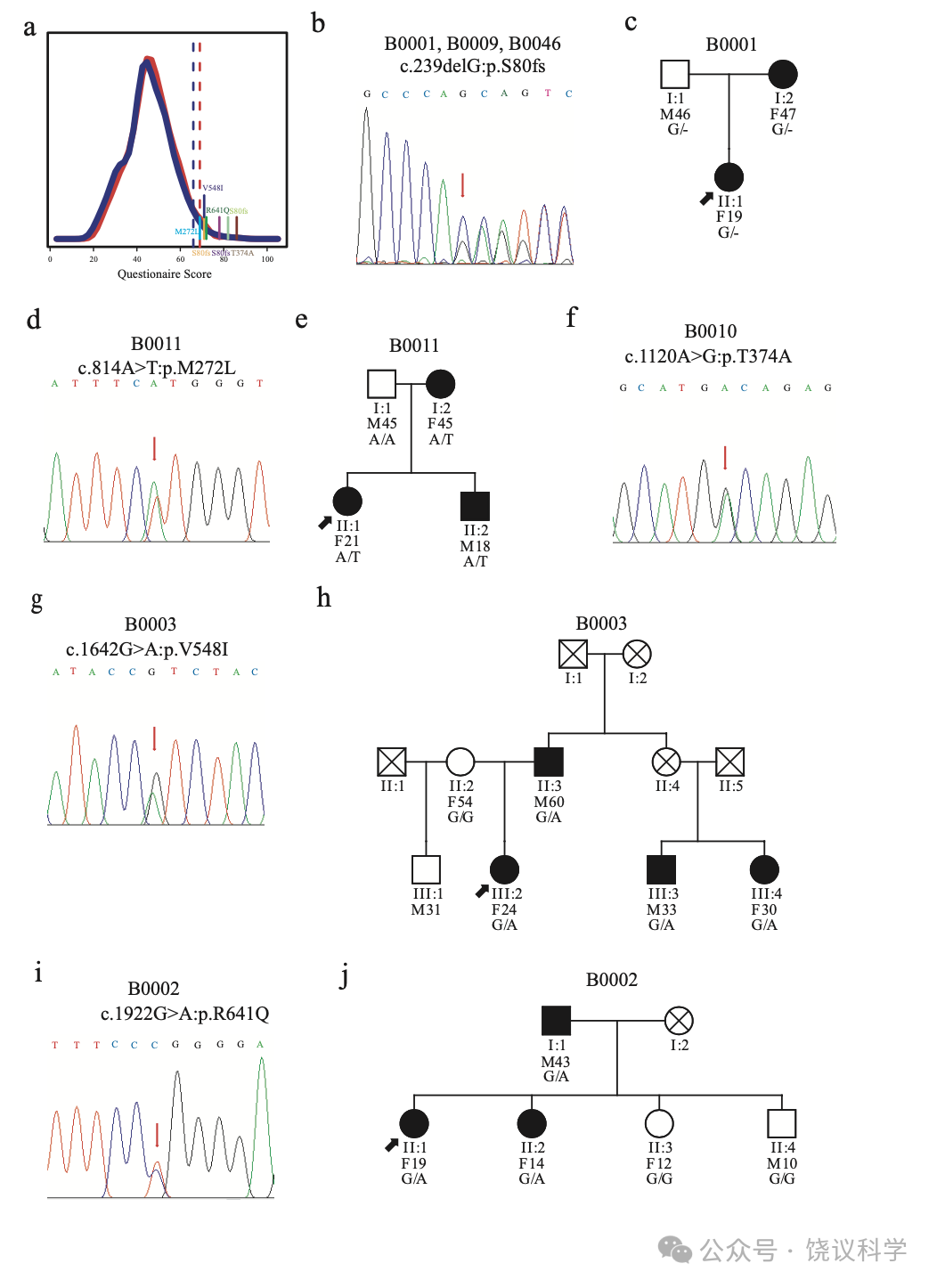
依据基因的相关分析揭示MCTP2罕见等位基因与脸识别的关联
下一步,我们在第二个有1928人的队列中也用问卷检查,得到脸识别能力,评估其与MCTP2等位基因对蛋白质编码有中度或重度影响的罕见变异的相关性。
基于负荷测验,在男性队列中,即使多次校正,检测到破坏性变异加全部异义突变有显著相关(所有变异pburden = 0.0009, poptical = 0.0021,更可能有害的突变pburden = 0.0032, poptical = 0.0063)。女性队列和同意变异中没有观察到相关性。表2显示贡献于这些显著性检测的变异的细节。我们的结果提示MCTP2基因高比例的因果变异往同一方向其作用,显示男性队列变异负荷与脸识别的相关性至少部分是因为的MCTP2作用。
CP个体中移码突变
家系和群体研究结果支持改变蛋白质编码的MCTP2罕见等位基因与脸识别能力相关,意指MCTP2参与脸识别。如上所述,2904位个体筛选到75位脸识别弱者中3位携带MCTP2基因第一个外显子的移码突变c.239delG(p.S80fs),这去除MCTP2蛋白质的大部分。这样的频率在已报到的CP发生率范围中,足够高到可以通过反向表型途径在同意分析脸识别的人中找更多的携带者。
我们进一步在第三个含1757人的独立队列找这一移码突变。我们检测到16位杂合子携带者MAF为0.0046。其中14人同意进一步检查(附表S3)。我们的面试记录了14位携带者与19位来自同一队列的非携带志愿者之间人才脸识别行为的定量差别(表3)。
四位(C2180、C2666、C3164和C3282)在我们联系之前就意识到其脸识别困难。检测C2666和C3164的全部能够检测的家庭成员显示这一突变在其两个家庭都与脸识别缺陷共分离(图3a和3b, 附表S3)。
C3049没有报告自己认人、认脸有可能,而是感到自己与其他人认人、认脸不同(附表S3)。C3049的家庭(图3c, 附表S3)内, 突变还见于两个亲戚(II:2 and III:1), 都有日常认脸困难。
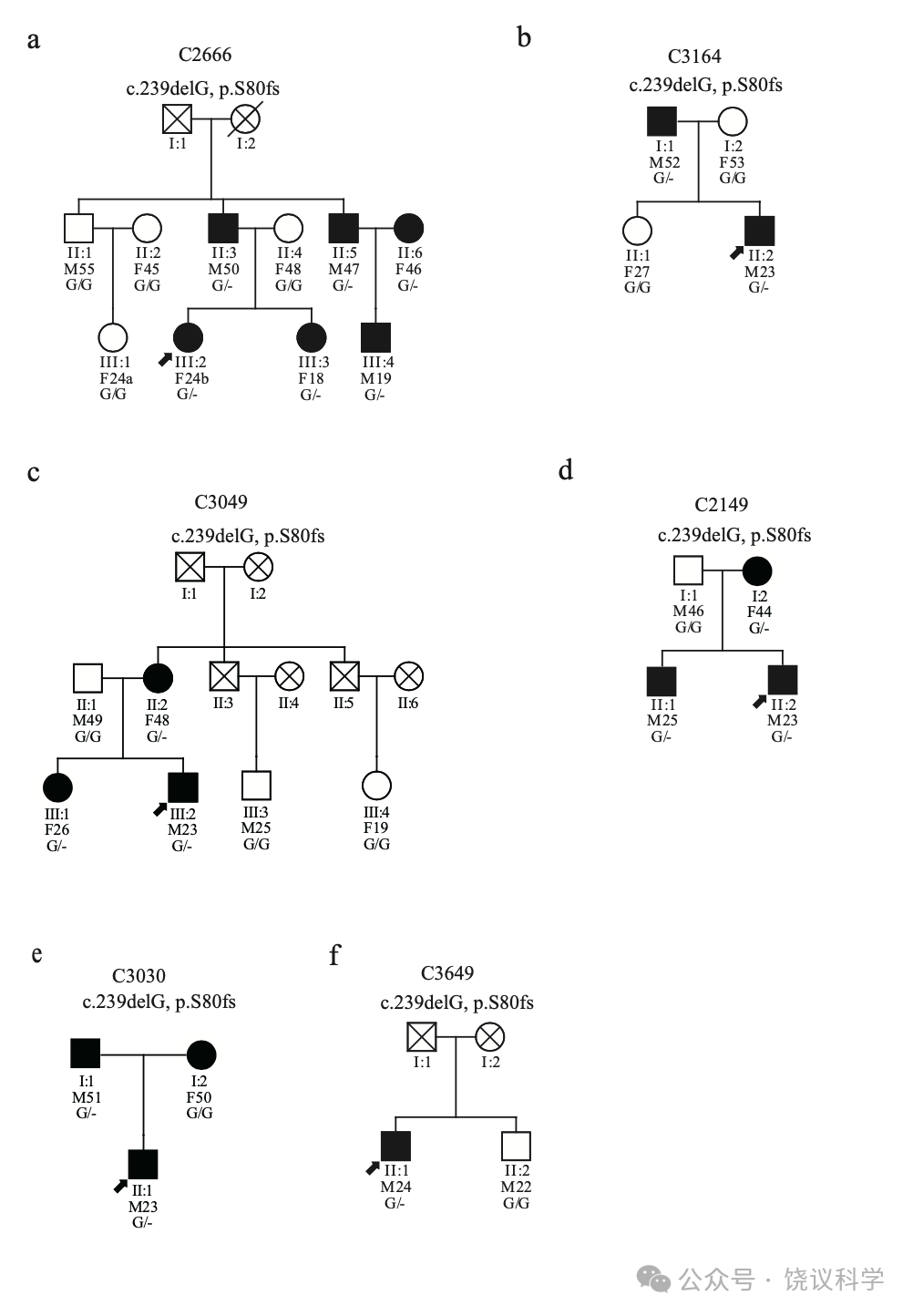
其余9位在面试中认为自己与其他人一样、甚至更能认脸(附表S3)。但是,其中8位(C2149、C2259、C3030、C3234、C3358、C3420、C3649 和C3731)发展了适应的行为,依赖明确的学习策略认脸,这样日常识别熟人(全人, 不局限脸)不难。当他们遇到没有特征的生人,屏幕上的演员(特别是女演员),或者熟人但脱离场景,策略就不会都很好地工作,但他们再调节、并很快更新信息。C2149的携带这一突变的亲戚抱怨识别困难(图3d, 附表S3)。C3030的认人的策略遗传自父母(图3e, 附表S3)。C3649的亲戚没有脸盲、也不携带这一突变(图3f, 附表S3)。C2147在整个面试中没有显示明显的异常脸识别。
通过收集母-父-后代三人的微卫星资料(附图S6),排除共同祖先来源的可能性,估计出c.239delG(p.S80fs)突变旁推测的单倍型。
MCTP2 突变CP家族神经成像研究
用A家族的6位成员,我们发现FFA区域对脸的异常反应与MCTP2 基因的I716S突变相关。
多多年的研究一直观察到rFFA参与检测脸的存在和区分不同的脸(Grill-Spector et al., 2004; Haxby et al., 2000; Kanwisher et al., 1997; Puce et al., 1996; Rangarajan et al., 2014; Rotshtein et al., 2005; Yovel and Kanwisher, 2004; Zhang et al., 2012)。正如在正常参试者的rFFAs(附图S7a), 有I716S突变(A-VI:5, 附图S7b, A-VI:7, 附图S7c与A-VI:9, 附图 S7d)和没有I716S突变(A-VI:1, 附图S7e, A-VI:2, 附图S7f 与AVI:10, 附图S7g)的家庭成员都有显著的rFFAs活动。这些提示,独立于突变,A家庭成员的rFFA都特异地对脸有反应。
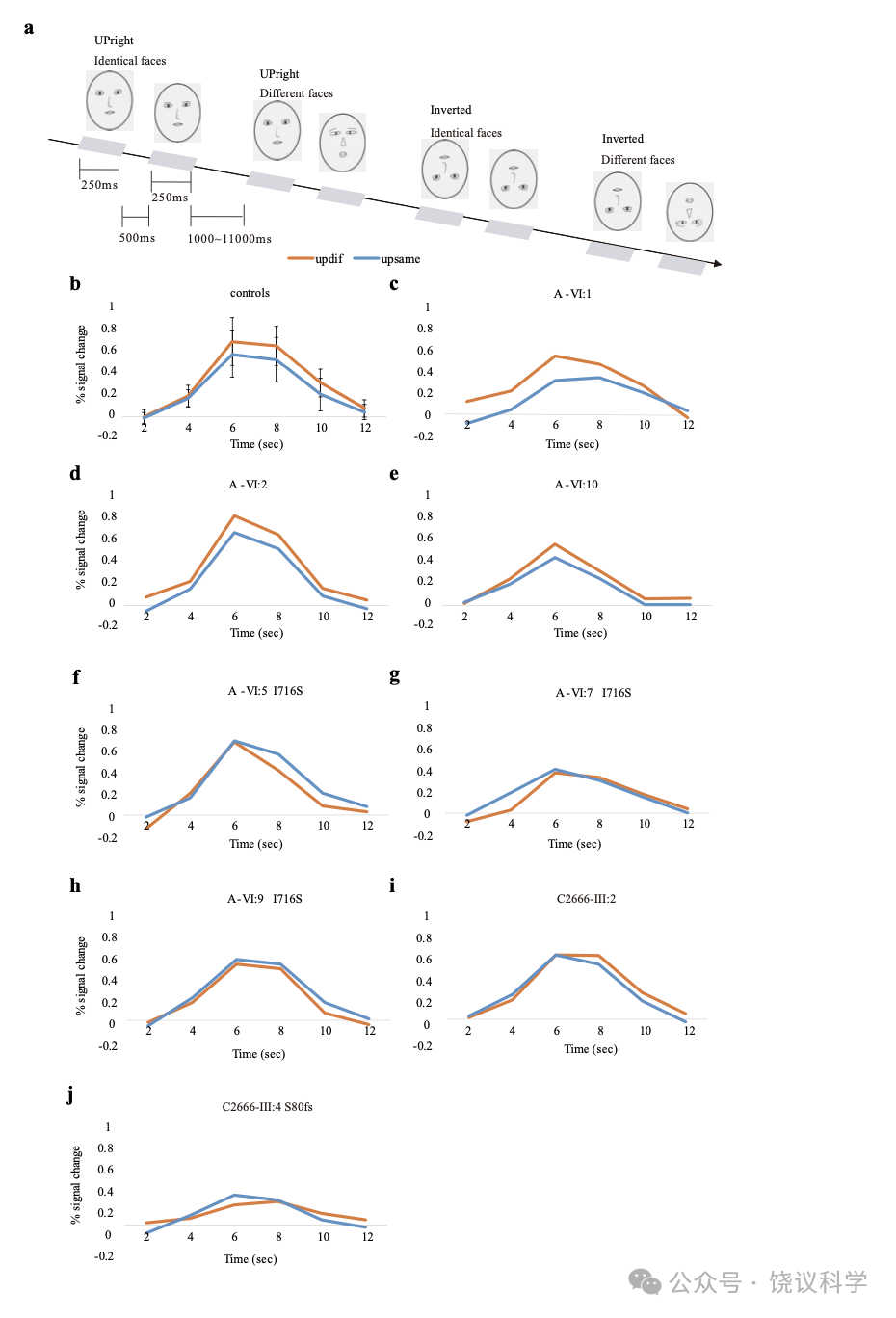
为了检测家庭成员的rFFA能否加工脸的个体差异,我们用事件相关设计fMRI适应模式进行实验(Yovel and Kanwisher, 2005)(图4a)。在正常组,fMRI对脸个体特征适应的可预计的重复压制在rFFA高度显著(配对 t-建议 p < 0.0001, 图4b)。而且,每单个受试者对一对正置的不同的脸(UDF)的引起的活动水平都高于一对正置的相同的脸(UIF):‘UDF>UIF’ 的差别在20位正常人的18人中呈p < 0.05的显著,并在其余受试者中在预计的方向有非显著的趋势(p < 0.057, p < 0.25)。在每一位没有I716S突变的家庭成员(A-VI:1, 图4c, A-VI:2, 图4d 与A-VI:10, 图4e),UDF对引起的活动高于UIF对。‘UDF>UIF’差别在每一位没有MCTP2突变的家庭成员呈p < 0.05的显著性。相反,有I716S突变的家庭成员(A-VI:5, 图4f, A-VI:7, 图4g and A-VI:9,图4h)之rFFA区域fMRI信号对不同脸的反应并不高于相同脸。有I716S突变的家庭成员的适应分(在感兴趣区域ROI,正置不同条件对正置相同条件百分比的变化)低于每一位正常对照受试者(图5),有I716S突变的家庭成员对比正常对照有相同方向缺乏显著的趋势(A-VI:5, p = 0.05407, A-VI:7, p = 0.12832 与A-VI:9, p = 0.06548)。因为重复同样脸而导致rFFA神经活动降低看起来也在有I716S突变的个体减少,而不同脸在CP、非CP家庭成员和正常对照引起类似的rFFA活动。这些结果提示MCTP2基因突变的家庭成员rFFA对相同脸的反应受损,这样可以有机制解释为什么识别已经见过的脸有困难。
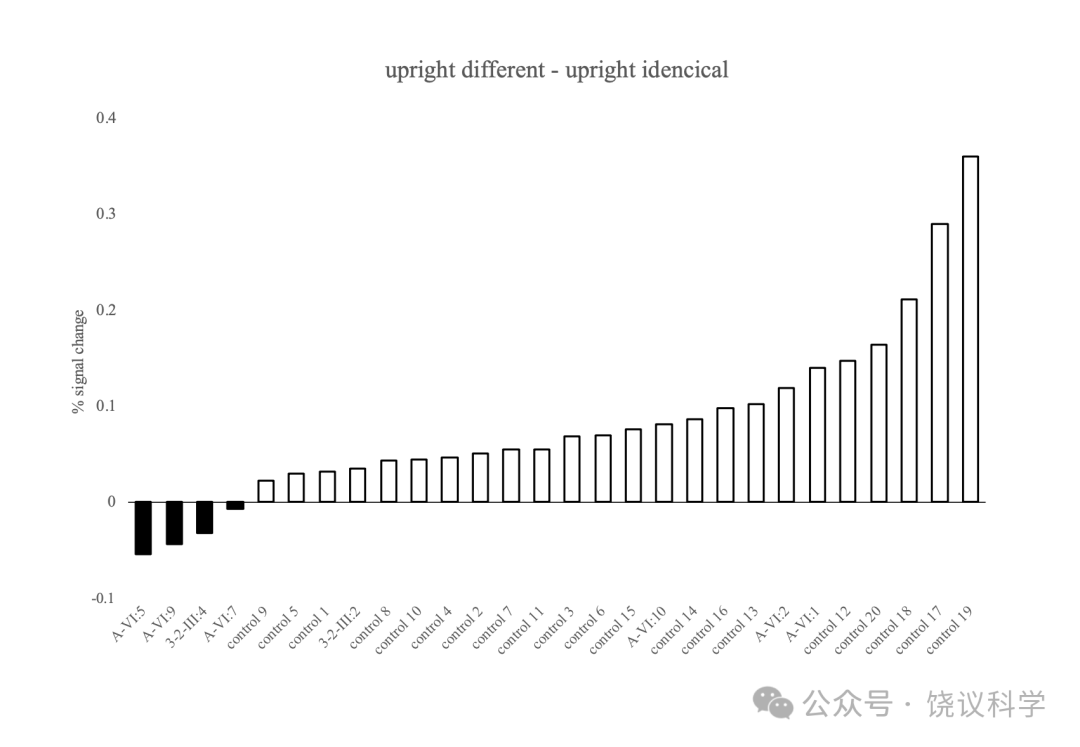
C2666家有MCTP2基因的c.239delG(p.S80fs)突变, 我们观察到p.S80fs的III:4(附图S7h)和无p.S80fs的III:2(附图S7i)的rFFA都对脸有正常反应,但在日常脸识别困难的个体III:4(图4j)的rFFA之fMRI对同样脸的重复也缺乏适应。
讨论
脸识别的分子遗传学研究
脸盲为人类认知(脸识别)的遗传分析模型。已经报道几个延伸的CP家系(Duchaine et al., 2007; Johnen et al., 2014; Schmalzl et al., 2008), 但其致病基因不清楚。我们的研究总体来说是对人类认知的第一个分子遗传学研究,具体来说是用延伸家庭对脸识别的第一项分子遗传学研究。
我们的发现MCTP2基因的突变对人类CP有贡献。这一结论依据与一下:1)连锁分析常染色体显性CP家庭A发现位于15q26.1-q26.2的CP位点(图1, 附图S3); 2)一个MCTP2 基因的特有突变c.2147T>G(p.I716S)是A家系中在MCR全基因组测序找到的唯一完全与CP共分离的可以改变蛋白质序列的突变(图1); 3)从2904人组成的队列中找到的75位脸识别困难的个体中7位脸盲者中发现MCTP2 基因5个罕见突变(Figure 2); 4)这7位脸盲者中,4位的家族成员愿被分析,皆显示基因型与表型的相关性(图2c, 2e, 2h与2j); 5)1757受试者的另一队列有16位携带与第4点里面3位脸盲个体相同移码缺失突变c.239delG(p.S80fs), 其中14愿被进一步分析。14位携带者日常脸识别行为不同于来自同一队列的19位非携带者(图3); 6)14位的4位有无疑的脸盲,其中两个家系愿被分析,结果都支持c.239delG(p.S80fs)与脸盲相关(图3a与3b), 进一步的支持来自更多家系(图3c、3d、3e与3f),他们发展了明确的策略用非脸的线索克服其脸识别困难; 7)在1928位组成的队列基因相关分析也检测到了MCTP2基因罕见等位基因与脸识别能力的相关性;8)在神经成像研究中,MCTP2基因突变的家族成员脸识别缺陷与rFFA对个体脸反应异常相关联。
CP的诊断
不同的认知任务研究CP显示异质性(Behrmann and Avidan, 2005; Kress and Daum, 2003; Le Grand et al., 2006; White and Burton, 2022)。虽然特异检测被证明有用(Duchaine and Nakayama, 2006a), 没有一个单独的检测有足够区分全部CP的能力(Duchaine and Nakayama, 2006a; Duchaine and Nakayama, 2004; Duchaine and Weidenfeld, 2003; Grueter et al., 2007; Shah et al., 2015)。
无论用哪个特定的任务,即使在同一个家庭类似的遗传和环境背景中(Johnen et al., 2014; Lee et al., 2010; Schmalzl et al., 2008),都需要谨慎行为检测个体CP的严重性受很多因素影响,例如行为代偿,其他认知技能,脸或检测范式的经历。在有些CP病例,他们识别人有困难,但他们在行为检测中却还能表现相当好。脸加工任务表现差可以预计脸识别缺陷,而相反不成立。
除了用行为检测诊断脸识别,也常常需要依赖个体自我报告。但是需要考虑Dunning-Kruger(DK)效应,一种元认知现象, 在某个任务表现差的人错误地自认为比其他人更好,而表现很好的人自认为不如其他人(Dunning et al., 2003; Kruger and Dunning, 1999)。DK效应见于脸识别自我报告(自我估计)和他人估计(同行估计)(Zhou and Jenkins, 2020)。理解脸识别自我报告检测时需要谨慎:很可能病例不知道其脸识别缺陷。但是通过仔细面试,可以发现他们有策略和日常习惯。同时,在DK效应中,虽然好的受试者低估其能量,他们只估计自己相当于平均,因此这不会显著影响其后表型判断,至少他们还被考虑为正常。因此,在我们的研究中,我们通过仔细面试家系成员作为首要的诊断基础。
MCTP2的作用
MCTP2基因编码的蛋白质有3个C2区域和两个跨膜区,类似参与突触传递的蛋白质(Shin et al., 2005)。其C2区域结合Ca2+(Shin et al., 2005)。很多有Ca2+结合C2 区域的蛋白质参与膜和囊泡转运, 在神经传递其关键作用(Cho and Stahelin, 2006; Shupliakov and Brodin, 2010)。
MCTP类蛋白质的C2和跨膜区在非脊椎动物就保守,如线虫和果蝇只有一个MCTP基因, 到哺乳类有两个基因(MCTP1 与MCTP2)。MCTP1表达于纸上神经系统,被意指参与中枢神经系统神经元和突触的内吞循环(Qiu et al., 2015)。在果蝇, MCTP参与稳定突触传递和自稳态可塑性(Genc et al., 2017)。斑马鱼的4个MCTP基因表达在神经和肌肉系统。 敲低MCTP2b i影响胚胎发育(Espino-Saldaña et al., 2020)。
人的MCTP2与MCTP1 基因都与高度遗传的神经精神疾病注意缺乏/多动症(ADHD)有关联(Kweon et al., 2018; Mick et al., 2010)。 “钙离子结合”的基因类别在14中ADHD相关基因中富集(Poelmans et al., 2011)。基因组分析在双极病找到MCTP1 单碱基多样性(SNP)(Scott et al., 2009)。也有一篇报道精神分裂症的MCTP2 SNP(Djurovic et al., 2009)。
在上面提到的1757人的第三个队列中,共找到并用Sanger测序验证了MAF < 0.005的64位编码或剪接位点罕见变异。在志愿同意的基础上进行了面试。16位个体携带 移码确实突变 c.239delG(p.S80fs)(MAF of 0.0046),14位接受了面试。在无关的S80fs变异携带者与脸识别能力直接维持了相关性。我们找到一个携带S80fs 家庭(C2666)有10位成员,突变与脸盲也有相关性。有趣的是,不仅携带S80fs 的II:5有脸盲,其妻子II:6也有同样突变并有脸识别困难。我们进一步面试了MCTP2基因其他低频突变的个体。还有更多的个体及其家庭含移码突变和剪接位点突变也显示脸识别异常。
从成千样本得到的基因表达微队列资料显示MCTP2基因表达于人脑, 包括颞叶(McCall et al., 2011)。被脸选择性一致可靠地激活的脑区FFA, 是颞叶皮层纺锤回的一个小区。人脑蛋白图谱显示了MTCP2蛋白质表达和分布概括,包括在纺锤回(Sjostedt et al., 2020)。
fMRI作为诊断内表型的方法并提供表型与脑活动之间的机理联系
fMRI显示特定任务/行为或条件下神经元激活区域。在复杂的神经行为异常疾病,内表型例如如fMRI被认为可以较临床检测类别更好地代表病理生理学(Rasetti and Weinberger, 2011)。神经成像-遗传读出允许更分层地区分特定危险等位基因对脑活动的影响,而不仅是简单诊断表型。
在本研究,我们在适应范式(Grill-Spector et al., 2006; Grill-Spector and Malach, 2001; Henson and Rugg, 2003)加入了成像-基因研究来显示家庭中脸识别困难成员受I716S或S80fs突变的影响。在正常对照者、以及家庭中没有MCTP2突变的成员中,在重复呈现同样的脸的时候,rFFA区域的神经信号显示降低,而在携带MCTP2突变的家庭成员中没有这种适应。
未来研究中, fMRI可以考虑为脸识别的重要内表型。
参与脸识别的基因
我们的遗传结果提供证据表明MCTP2基因突变导致先天脸盲,我们fMRI结果提示MCTP2基因参与区分脸所需要的神经环路。
当然,高级认知需要很多细胞和分子。MCTP2不会是唯一参与CP的基因。从2904人队列找到的75位CP患者中只有7位携带MCTP2基因突变。从1928人队列中女性脸识别能力与MCTP2罕见等位基因没有相关性。更多的连锁研究将有帮助。
我们发现更多家庭没有遗传基础,这需要更多分析。无假设的基因组分析可以考虑用下代基因组分析,包括常见和罕见遗传变异,需要大样本,超过百万人,找到更多遗传线索,也验证我们有关人脸识别的MCTP2 结果。
材料与方法
受试者和方法
受试者细节
本研究为北京大学保护人类和动物受试者委员会所同意。试验前,参与者或其法定代表人书面知情同意参与研究并允许发表研究结果。基因组DNA用Gentra Puregene Blood Kit(QIAGEN)从全血whole blood(BD Vacutainer 3.2% Sodium Citrate(1:9))获得、或用GeneFiX DNA Saliva Collector and Isolation Kit从唾液获得。
家系A
A家系的先证者V:11(图1a, 表1)来自自我介绍。用标准的半结构化面试(Carbon et al., 2007; Grueter et al., 2007; Kennerknecht, 2021; Kennerknecht et al., 2006)评估了该家系的35位个体(18 CP含9男9位,年龄16至72岁; 17位非CP, 9男8女, 年龄15至78岁)。25位年龄低于60的继续检测用了剑桥脸记忆检测-中文(Cambridge Face Memory Test-Chinese, CFMT-C)(Bowles et al., 2009; McKone et al., 2012), 配对剑桥车记忆检测(the matched Cambridge Car Memory Test,CCMT)(Dennett et al., 2012)与脸倒置效应(the Face Inversion Effect,FIE)区别检测(Yovel and Kanwisher, 2005)。 34 individuals(not including VI:9, who contacted us voluntarily after we had finished genotyping)were included in the linkage study. Nine individuals(V:1, V:4, V:6, V:9, V:11, V:13, V:15, V:19 and VI:8)were selected for WGS. This information is listed in Table S1. None of the persons showed signs of an autism spectrum disorder or neurodegenerative disorders.
第一个2904人队列中筛选到的更多有MCTP2 突变的CP患者
To investigate more CP individuals and examine whether rare functional mutations inMCTP2 are present in more CPs, the first cohort of 2904 individuals(average age of 19.25 + 1.30, 2161 females, 743 males, Jiangxi province )from the Westlake BioBank for Chinese(WBBC)pilot project(Zhu et al. 2021a; Cong et al. 2022a; Cong et al. 2022b), were screened with a questionnaire adapted from the 20-item self-report measure(see CP Questionnaire in Methods Details)for quantifying CP traits. 78 individuals scored worse than the mean by 2 SDs(Figure 2a). 75 of them provided DNA samples. 44 individuals including 7 individuals with MCTP2 mutations agreed to an interview for CP diagnosis and family members were contacted for availability for further studies.
第二个1928人队列的基于基因的相关分析
The second cohort of 1928 individuals(average age of 18.51+ 0.93, 1085 females, 843 males, Jiangxi province)used in the gene based association of the rare functional alleles in MCTP2 with face recognition ability were also from the WBBC project(Zhu et al. 2021a; Cong et al. 2022a; Cong et al. 2022b). The coding sequences of the MCTP2 gene for each person was analyzed by tagged-amplicon deep sequencing.
从第三个含1757人队列筛到的MCTP2移码突变c.239delG(p.S80fs)个体
A third cohort of 1757 individuals(average age of 19.13+ 1.07, 1295 females, 462 males, Guangdong province)were sequenced for the presence of the c.239delG(p.S80fs)mutation in the exons of MCTP2. The coding sequences of the MCTP2 gene for each person was analyzed by tagged-amplicon deep sequencing. For individuals carrying p.S80fs, their family members were further contacted to test for the S80fs mutation by direct Sanger sequencing and assessed by the standardized semi-structured interview. The differences in the behavior of daily face recognition between the 14 carriers and 19 non-carrying volunteers from the same cohort were analyzed by independent sample t-test.
行为检测的对照样本
338 normal participants(average age of 42.68+ 16.53, 164 females, 174 males)were tested. These participants were unrelated. Participants were not selected for face recognition ability with no known history of major brain injury, or other major disorders likely to affect face recognition(e.g., Alzheimer’s disease), representing a random sample of the community. They were tested on a battery of tests including the CFMT-C, the CCMT, the FIE Test and the Cambridge Face Perception Test - Chinese(CFPT-C).
fMRI受试者
Age-related dedifferentiation and compensatory changes in the functional network underlying face processing have been found in studies(Burianova et al., 2013; Germine et al., 2011; Goh et al., 2010; Le Grand et al., 2006; Lee et al., 2011; Park et al., 2012; Zebrowitz et al., 2016). Therefore, in order to exclude age influence on fMRI results, we mainly studied family members younger than 30 years old. Family members who participated in the fMRI experiment were on a voluntary basis. In family A, 3 MCTP2 mutant CP individuals(VI:5, VI:7 and VI:9)and 3 non-mutant and non-CP individuals(VI:1, VI:2 and VI:10)took part in fMRI; in family 3-2, III:4(CP, with S80fs)and III:2(non-CP, without S80fs)took part in fMRI.
21 non-CP students(average age = 23.63+ 3.71, 6 females, 15 males)were recruited for fMRI analysis.
CP referred to those with obvious face recognition problems in daily life diagnosed by the structured interview.
All participants reported normal or corrected to normal vision, no history of neurological or psychiatric conditions and all were right-handed. Anatomical volumes(i.e., structural MRIs)had been routinely checked. One of the normal students was excluded from neuroimaging studies because the maximum head rotation was over 1.5 degree or the maximum translation was over 2 mm during localization.
We adopted the methods of studying single-cases that the severity of each individual could be reported by comparison with the control population(Bentin et al., 2007; Busigny and Rossion, 2010; Dricot et al., 2008; Gao et al., 2019; Hadjikhani and de Gelder, 2002; Righart et al., 2009; Rossion et al., 2011; Schiltz et al., 2006). In fMRI analysis, each family member was compared individually to a small sample of normal controls by a modified t-test(Crawford and Garthwaite, 2002; Crawford et al., 2010; Crawford and Howell, 1998).
方法细节
CP诊断的结构面试
The diagnosis of CP was based on a standardized semi-structured interview(Tables 1, S2 and S3)(Grueter et al., 2007; Kennerknecht et al., 2006),加一个Grueter2008 which documented a variety of impressive qualitative differences between CP and non-CP, and had been validated with objective face recognition tests in previous studies(Carbon et al., 2007; Grueter et al., 2007). These criteria were employed in recent literature on CP(Bate et al., 2019; Bate et al., 2014; Carbon et al., 2010; Dinkelacker et al., 2011; Esins et al., 2016; Esins et al., 2015; Gruter et al., 2009; Kennerknecht et al., 2008b; Stollhoff et al., 2010, 2011; Verfaillie et al., 2014; von Kriegstein et al., 2008; Zhao et al., 2018; Zhao et al., 2016).
The interviewer asked questions in a semi-structured interview format with three or four questions about each diagnostic item. Interviews were held to embed the questions into conversations and questions about the same diagnostic items not asked sequentially. Interviews included a medical history in order to exclude conditions which might cause or mimic prosopagnosia. Affected people present a lack of confidence with face recognition. Symptoms include lasting and irritating subjective uncertainties of face recognition, failure to recognize familiar people out of context or in crowded places, overlooking familiar people and confusing strangers with familiar people, face recognition/learning time longer than socially accepted, development of adaptive behavior of critical situations and strategies for visual recognition of people and time of onset.
Consistent with the interview results, some individuals were aware of their CP before we contacted them. Some individuals who identify people via non-facial features such as voice, gait and general appearance and manner, or were unaware of face recognition problems, but had developed obvious compensatory strategies to cope with difficulties were also diagnosed as CPs.
CP问卷
To effectively screen CP candidates from big samples, we adapted a Chinese 20-item version from the 20-item self-report measure for quantifying prosopagnosic traits(Shah et al., 2015), which asks about tangible experiences. This 20-item Questionnaire was included in a set of questionnaires for many research purposes and filled out by two cohorts of individuals online. Invalid questionnaires were dropped due to no distinction between forward and reverse questions. 2904 valid questionnaires for the first cohort and 1928 valid questionnaires for the second cohort were collected. The internal reliability measured by Cronbach's α was 0.828 and 0.902 respectively. In the first cohort, 343 individuals finished the questionnaire for a second time several weeks later and the correlation for the first and second results across each individual was very high by Pearson correlation coefficient analysis, r = 0.081, p < 0.001. There was a significant difference of the score distribution between the female and male participants(p < 0.001, two tailed t test), so the candidate CPs were screened and the gene based association study were carried with different genders respectively. An individual with a score above 2 SDs of the mean of the controls was defined as a CP candidate.
行为检测的刺激和程序
All behavioral tests were adapted and integrated into a whole set using the Hyper Text Markup Language. All participants were tested individually. The tests were run on a desktop PC with screen resolution 1024 × 768, refresh rate 85Hz. Participants were seated at a viewing distance of approximately 50 cm from the screen. All participants were tested wearing their usual optical correction. Participants were asked to confirm that they could focus without seeing blur on the computer screen. No participant reported any difficulty with focus at these distances. Gray-scale adult Chinese faces were used.
剑桥脸记忆检测-中文(CFMT-C)
The Chinese face version of CFMT(McKone et al., 2012)was kindly provided by Professor Jia Liu of Beijing Normal University which we integrated with other behavioral tests into a whole set, and performed according to the standard procedure including the practice phase, the “Learn” phase with 18 trials, the “Novel Images” phase with 30 trials and the final “Novel Images with Noise” with 24 trials. All faces were Chinese male, shown without hair or facial blemishes and with neutral expressions. Participants were instructed to press the key corresponding to the number 1, 2 and 3 below faces. The test includes a total of 72 trials. Scores were reported as percent correct across the full test.
剑桥车记忆检测(CCMT)
The CCMT is a test similar in the experimental design as CFMT, with stimuli replaced by whole cars(Dennett et al., 2012). We used the CCMT as a control of the CFMT to test for potential general object recognition deficits and to quantify the individual ability of performing such kind of tests. The original version of CCMT was kindly provided by Professor Bradley Duchaine of Dartmouth College, USA, which we integrated with other behavioral tests into a whole set, and performed according to the standard procedure(Dennett et al., 2012)
剑桥脸感知检测-中文(CFPT-C)
We developed a version of CFPT using Chinese faces as stimuli(CFPT-C), according to the standard procedure(Duchaine et al., 2007)by morphing six different individuals with the target face, containing 88%, 76%, 64%, 52%, 40%, and 28% of the target face in turn. The Chinese faces were male, shown without hair or facial blemishes and with neutral expressions. All faces were photographs of Chinese students at Peking University, with written consent forms collected before photographing. Each individual was photographed in the same range of views and lighting conditions. Eight upright and eight inverted trials were intermixed, with the upright trial occurring first half the time. Participants had one minute to arrange six morphed faces according to their similarity to a target face by clicking on a face and indicating where that face should be moved by clicking in the area between two faces. Scores were computed according to the previous paper(Duchaine et al., 2007). The internal reliability measured by Cronbach's α in our sample of 170 individuals was 0.705 for upright faces, and 0.483 for inverted faces. However, after our pilot study, the CFPT-C was excluded from further study, because strategies such as just comparing partial facial features were used in normal people as in CPs and inconsistent results were reported by others(Anstey et al., 2005; Bowles et al., 2009; Duchaine et al., 2007).
脸倒置效应(FIE)区分检测
The stimuli consisted of 20 gray-scale individual face images, cropped using the same 4 X 4.5 cm oval window(4.6º X 5.2º of visual angle)to remove cues from the hairline and face contour. All faces were photographs of Chinese students at Peking University, with written consent forms collected before photographing. Photographs were not repeatedly used in different tests. Pairs of face stimuli were presented sequentially either upright or inverted in a randomized order. The first face stimulus was presented in the upper-left quadrant of the screen for 0.5 s. After an interstimulus interval of 0.5 s, the second stimulus was presented in the lower-right quadrant for 0.5 s. The next trial will not begin until a same or different response was made by pressing one of two keys to respond by the participant. 80 trials were conducted in this test, half of them with upright faces and half with inverted faces. In each condition(upright or inverted), the chance with the two identical faces was 50%. A few practice trials were presented before the beginning of the experiment. Scores were reported as percent correct for each condition. For the measure of the FIE, the difference in performance level between upright and inverted faces, an FIE index was calculated by entering the correction of performance for upright and for inverted faces in the following formula:FIE =(upright –inverted)/(upright + inverted).
行为检测的资料分析
Due to possible effects of ageing and sex difference on the scores of the CFMT-C, CCMT, upright faces and inverted faces in the FIE and the FIE index, we used multiple stepwise regression analyses to identify the covariates(sex, age)specific to each trait from 338 participants(aged 15–83 years, 164 females, 174 males). The results of the CFMT-C and the CCMT showed noticeable age-related decline and sex differences(Figure S1). The scores of upright faces and inverted faces in the FIE correlated with ageing but not sex(Figure S2), while the FIE index was not affected by age or sex among the control samples.
Examining function curves suggested that the behavior performance remained stable across early middle age, but began to decline noticeably at approximately 50 years of age. This is consistent with previous studies. In terms of the validity of the behavioral scores, only individuals with ages under 60(average age = 36.51+ 12.34, 130 females, 138 males)were included to calculate the best estimate of CP cut-offs.
We used the fit-and-residual procedure to calculate the standard residual of each participant(Z score)for CP family members as described previously(Bowles et al., 2009). The regression function describing the relationship between age, sex and CFMT was:CFMT score = -0.212 age + 6.188 sex + 73.724. The standard deviation(SD)of the controls’ residuals of CFMT score was 11.189. The regression function for CCMT was:CCMT score = -0.134 age - 2.968 sex + 69.444, and SD was 10.252. The regression function for upright faces was:Upright score = -0.170 age + 87.641, and SD was 8.862. Z score was calculated by dividing the participant’s residual by the SD of the controls’ residuals.
For the FIE index, there was no correlation with age or sex. Normalized Z score was calculated for each subject by subtracting the mean of the control sample and dividing by the control samples’ SD.
遗传分析
连锁分析的全基因组基因分型
DNA samples were genotyped using Infinium Human OmniZhongHua BeadChips(Illumina), and normalized bead intensity data obtained for each sample were converted into SNP genotypes using Genome Studio. SNPs were then selected according to the following parameters:genotyping rate > 95%, minor allelic frequency > 1% and no significant deviation from Hardy–Weinberg proportions(p> 0.001)by PLINK software(Purcell et al., 2007). Gender corresponding to each DNA sample was checked by analysis of X chromosome heterozygosity using PLINK. The initial Mendelian inheritance in family A was analyzed by PLINK and KING toolset(Manichaikul et al., 2010).
连锁研究的资料分析
For the preliminary linkage analysis in Family A(named linkage 1)(Table S1), we diagnosed 9 family members as CP cases(V:1, V:4, V:6, V:9, V:11, V:13, V:15, V:19 and VI:8)with the criteria of not only abnormality in face recognition exhibited through interviews, but also poor with the CFMT behavior test(under -1.5 SDs). 11 family members with normal daily face recognition by interview and Z scores of all the behavior tests over -1.5 SDs were taken as normal controls(V:3, V:7, V:8, V:12, V:14, V:16, VI:1, VI:2, VI:3, VI:4 and VI:10). The remaining subjects in the pedigree were set as unknown at this stage of analysis, including those over 60 and not suitable for behavioral tests.
We did a second linkage analysis(linkage 2)after adding four individuals who had daily face recognition problems but appeared normal on behavioral tests(V:10, VI:5, VI:6 and VI:7)and four founders with daily face recognition problems(IV:2, IV:4, IV:6 and IV:10)as CPs(Table S1), and the remaining six founders without daily face recognition problems(IV:3, IV:5, IV:7, IV:8, IV:9 and IV:11)as controls. They were set as unclear in the linkage analysis analyzed above(linkage 1).
Parametric linkage analysis was performed with the Merlin programs(Abecasis et al. 2002), assuming autosomal dominant inheritance with 100% penetrance, disease allele frequency 0.001, and phenocopy rate 0.05. Because the presence of linkage disequilibrium(LD)might inflate linkage statistics, LD maps were constructed with the PLINK tool within family members with the LD thresholds(r2 < 0.5). Following data QC, 177,126(r2 < 0.5)informative SNPs were selected for linkage analysis with Merlin(Abecasis et al. 2002). 1000 simulation analyses were performed to exclude false positive results due to random chances under the null hypothesis of no linkage, with simulated data, while maintaining the pedigree structure, allele frequencies, and recombination fraction. For positive regions, haplotypes were constructed and subsequently checked manually on the basis of the minimal number of recombination.
拷贝数变异(CNVs)呼叫的生成
CNVs were identified for each member. Genotyping and signal intensity data were exported from the GenomeStudio software 2011.1(San Diego, CA, USA). The subsequent CNV calling analyses were performed using PennCNV(v.2011 Jun16)(Wang et al., 2007)according to the manual. No correlation was detected between the CNV genotypes and the phenotypes for each linkage analysis.
全基因组测序(WGS)
WGS was performed by the Next-Generation Sequencing Center of Biomedical Pioneering Innovation Center, PKU. Sequencing libraries were built with NEBNext Ultra DNA Library Prep Kit for Illumina.150 bp paired-end sequencing was done on Illumina HiSeq 4000.
WGS的资料分析
FastQC(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)was applied to perform quality checks on various aspects of sequencing quality. Low quality bases were removed by Trimmomatic software(Bolger et al. 2014)with the parameters HEADCROP:5, LEADING:25, TRAILING:25, SLIDINGWINDOW:4:15 MINLEN:35. After that, clean sequences were aligned to human genome build hg19 and SNPs, INDELs or structural variants were called by SPEEDSEQ(Chiang et al. 2015). BAM files locally realigned around INDELs were used to determine the average coverage, using GATK DepthOfCoverage package and default settings(McKenna et al. 2010). After the alignment and variant calling, the mean depth of all samples was 34, and 96% of the mapped bases were covered at more than 10-fold on average(Table S4). In the linkage region, 100% coding bases were covered at more than 20-fold in at least one of the four individuals. All variants were annotated to RefSeq hg19 and ANNOVAR(Wang et al. 2010)were used to add alternative allele frequencies, variant effect predictions and functional annotations. We excluded variants with minor allele frequency(MAF)> 0.05 in multiple databases, including the dbSNP(v150)(Sherry et al. 2001), the 1000 Genomes Project(1000g2015aug)(Genomes Project et al. 2010; Genomes Project et al. 2012; Genomes Project et al. 2015), the Exome Aggregation Consortium(ExAC03)(Lek et al. 2016)and the Genome Aggregation(gnomAD, v2.1.1, http://gnomad.broadinstitute.org)databases.
标记放大子的深度测序
Target-specific primers forthe coding sequences and the exon-intron boundaries of the MCTP2 gene(GenBank:NM_018349)were designed with universal primer sequences(termed CS1 and CS2)appended at the 5’-end and sequencing was performed as described previously(Forshew et al. 2012).
标记放大子深度测序的资料分析
The raw paired 150 bp-long reads were mapped to the human reference genome(build hg19)using BWA. GATK(McKenna et al. 2010)was then used to perform local realignment and recalibrate base quality scores, producing a BAM file for each individual. All variants were annotated to RefSeq hg19 and ten algorithms(Sift, Polyphen2_HDIV and HVAR, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, MutPred and VEST)integrated by ANNOVAR(Wang et al., 2010)were used to add alternative allele frequencies, variant effect predictions and functional annotations. Among these 10 algorithms, if a site was predicted as deleterious by Sift, LRT, FATHMM, or ROVEAN, as probably or possible damaging by Polyphen2HDIV or HVAR, as disease_causing_automatic or disease_causing by Mutation Taster, as high or medium by Mutation Assessor, and with scores greater than 0.5 by MutPred or VEST, it could be considered harmful for the specific algorithm. 91% of targeted bases were covered at ≥ 100-fold per individual(Table S4). We manually inspected each mutation using the Integrative Genomics Viewer(Robinson et al. 2011; Thorvaldsdottir et al.2013)to rule out false positive findings. If the coverage for each exon was under 100-fold depth in each individual, we performed direct Sanger sequencing to verify these regions.
在1928人组成的第二队列对MCTP2进行的基于单基因的负担分析
We resequenced all 22 exons, 5’UTR, 3’UTR and the exon-intron boundaries ofMCTP2 by the tagged-amplicon deep sequencing and did a single gene-based burden test for overall, female and male cohorts using the unified optimal sequence kernel association test(SKAT-O)(Lee et al. 2012). A total of 54 rare coding, four splice variants and 19 synonymous variants in NM_018349.4 with MAF ≤ 0.005 in EAS of 1000G, ExAC, and gnomAD were identified and verified by Sanger sequencing(Table S5). Primary analysestested 1)all variants:disruptive variants(nonsense, essential splice site and frameshifts)plus all missense variants(154 individuals carrying); 2)more likely to be harmful:disruptive variantsplus missense variants predicted to be harmful by at least five algorithms(79 individuals carrying); and 3)synonymous variants(35 individuals, an individual with both a synonymous mutation and a mutation from 1)considered as a 1)carrier ).
测序验证
Mutations were amplified by PCR and validated by direct Sanger DNA sequencing. All reactions were 100% successfully validated. Primer sequences for PCR amplification are included in Table S6.
Founder Origin Testing
We genotyped nine multi-allelic microsatellite markers(CHLC.ATA22D04, AFMB077YD5, GATA128A02, AFM072YB11, AFM357TD9/D15S1038, CHLC.GATA73F01, GATA161C02, AFM217ZG1, AFM309VG9)around the c.239delG(p.S80fs)mutation ofMCTP2 from 94255659 bp to 96210769 bp on chromosome 15 in families B0001, C2149, C2666, C3030, C3049 and C3164. The inferred haplotypes were estimated by comparison to these microsatellites data collected on mother-father-offspring trios to test the possibility of the same founder origin.
神经成像
刺激.
In the localizer experiment, images of faces, non-face objects(e.g., chairs, food, and tools), and texture patterns(scrambled faces)were presented at the center of the screen, subtended 6.2°×6.2°. In the adaptation paradigm, the stimuli were gray-scale images of young Chinese men(hair cropped with neutral expressions). The stimuli were presented by a MRI compatible projector system(SA-9900, The Shenzhen Sinorad Medical Electronics Co., Ltd, China, http://www.sinorad.com), with a spatial resolution of 1,024×768 and a refresh rate of 60 Hz.
实验设计和程序.
Each participant completed the same single scan session consisting of one functional localizer run and five runs for the adaptation paradigm. During the experiment, participants lay on their back in the scanner, using earplugs to reduce noise and sponges to hold their heads in place to reduce head movement. Participants viewed the stimuli presented on a translucent screen visible via a mirror mounted to the head coil at a distance of 60 cm. In the localizer run which lasted 360 seconds(s), with a 12 s dummy at the beginning of the run, images appeared at a rate of 2 Hz in blocks of 12 s, interleaved with 12 s blank blocks. There were five blocks for each type of images in the run. Each image was presented for 300 milliseconds(ms), followed by a 200 ms blank interval. Subjects performed a one-back task during scanning to ensure maintenance of attention to the stimuli. In the adaptation paradigm, pairs of face or house stimuli were presented sequentially either in the upright or the inverted manner in a randomized order that was optimized for the extraction of the hemodynamic response in an event-related fast presentation design. Each trial lasted 2000 ms. The first and second stimuli were presented for 250 ms, with an interval of 500 ms and followed by 1 s fixation. Blank screen with white cross fixation point was set between trials, with a presentation time of a random even number in the range of 0-10 s to optimize the efficiency of the event-related fMRI design by optseq2(http://surfer.nmr.mgh.harvard.edu/optseq). Each run lasted 332 s, containing eight conditions(upright same face, upright different face, inverted same face, inverted different face, upright same house, upright different house, inverted same house and inverted different house). Five runs were included, each containing 12 trials for each condition. Subjects made a same/different response on each trial. Here we focus only on the face stimuli.
fMRI扫描
The fMRI data were collected in a 3T GE MR 750 scanner, with an 8-channel phase-array head coil(GE Healthcare, Waukesha, WI)at Peking University Center for MRI Research.
The gradient-echo echo planar imaging(EPI)sequence was employed for the blood oxygenation level-dependent(BOLD)signal images acquisition, and the imaging parameters were as below:repetition time(TR)= 2000 ms, echo time(TE)= 30 ms, field of view(FOV)= 224mm × 224mm, matrix = 64 × 64, flip angle = 90°, slice thickness = 3.5 mm with 0.7 mm spacing, voxel size = 3.5 × 3.5 ×(3.5 + 0.7)mm, 33 oblique slices covering the whole brain).
The structure images were acquired by a 3D inversion recovery-prepped T1-weighted sequence(fSPGR, sagittal acquisition, TR = 6.65 ms, TE = 2.92 ms, TI = 450 ms, flip angle = 12°, FOV = 256 mm × 256 mm,matrix = 256 × 256, 192 continue slices with 1mm slice thickness, voxel size = 1 mm × 1 mm × 1 mm).
神经成像的资料分析
Preprocessing and data analysis were performed with SPM12(Wellcome Trust Centre for Neuroimaging, London; http://www.fil.ion.ucl.ac.uk/spm/software/spm12/). Functional images were sequentially processedin accordance with the standard SPM approach as follows:interpolated to correct for slice timing, realigned to the middle volume, co-registered to structural scans using the mean functional image, spatially normalized to a standard echo-planar image(EPI)template based on the Montreal Neurological Institute reference brain template(MNI152, Asia brain), and spatially smoothed with an isotropic 8 mm full width at half-maximum Gaussian kernel. For anatomical reference, the statistical maps computed were overlaid to the 3D T1-weighted scans. First, face-selective regions were localized in each subject by BOLD signals. By comparing the “faces” condition with the “non-face” condition in the localizer experiment at the first-level analysis, we assessed the face-selective region of interest(ROI)for every participant by building a 5-mm radius sphere surrounding the coordinate of the maximum activation in rFFA at a threshold p < 0.05, the family-wise error(FWE)correcting for multiple testing, with MARSBAR followed by visual confirmation of their anatomical location. For participants who lacked a face-selective area with this criterion, we explored liberal uncorrected significance thresholds(as liberal as p < 0.05), to avoid missing effects that might be apparent at less stringent uncorrected thresholds. Because faces are processed more dominantly in the right hemisphere and because the ROIs in this study were localized more consistently in the right hemisphere, we chose to restrict our ROI-based analyses to regions in the right hemisphere(unless noted otherwise). Second, the above-defined ROI were tested for the adaptation to facial identity using the contrast(up different faces > up identical faces)by BOLD signals at the first-level analysis. Third, the time course of percent signal change from baseline fixation was extracted from the ROI for each condition in each individual with MARSBAR, and was plotted for each condition in controls(+SD)and family members by an in-house Matlab program. Fourth, in order to directly compare each family member with the control subjects, the percent signal change in the ROI was computed for each condition. Three data points around the peak of the hemodynamic response defined individually were averaged to estimate the percent signal change. Fifth, the percent signal change was used to compute the adaptation scores(upright different face – upright identical face)for each subject, allowing a comparison between each family member with the control group by means of Z-scores and the modified t-test score Crawford and Howell, 1998(Crawford and Garthwaite, 2002; Crawford et al., 2010).
Results were visualized using xjView toolbox(http://www.alivelearn.net/xjview)in addition to built-in visualization in SPM12.
定量和统计分析
Where applicable, statistical parameters including sample size, precision measures(standard error or SD)and statistical significance are reported in the figures and corresponding legends.P-values of less than 0.05 are significant.
致谢
我们感谢参与此研究的所有人(特别是先证者及其家庭A), 感谢北京师范大学的刘嘉博士与我们分享剑桥脸识别测试的中文版、以及有益的讨论,感谢Darmouth学院的Bradley Duchaine博士贡献剑桥汽车记忆测试和有益的讨论,感谢北京大学的Jurg Ott、韩世辉和Taoyu Wu博士的有益讨论,感谢西湖大学的Pianpian Zhao博士、 上饶医专的Jing Jiang和Kunjun Mao 博士, 南方医科大学的Liang Chen和Jieyu Chen博士帮助收集样本, 感谢金子兵博士和张学博士对本文的批判性评论,感谢北大清华生命科学联合中心高性能计算平台提供计算支持,感谢首都医学科学创新中心、北京脑科学研究所、北大清华生命科学联合中心和深圳湾实验室的支持。
参考文献
Abecasis, G.R., Cherny, S.S., Cookson, W.O., and Cardon, L.R.(2002). Merlin[mdash]rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30, 97-101.
Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P., Kondrashov, A.S., and Sunyaev, S.R.(2010). A method and server for predicting damaging missense mutations. Nat Methods 7, 248-249.
Afraz, S.R., Kiani, R., and Esteky, H.(2006). Microstimulation of inferotemporal cortex influences face categorization. Nature 442, 692-695.
Allison, T., Puce, A., Spencer, D.D., and McCarthy, G.(1999). Electrophysiological studies of human face perception. I:Potentials generated in occipitotemporal cortex by face and non-face stimuli. Cereb Cortex 9, 415-430.
Anstey, K.J., Dear, K., Christensen, H., and Jorm, A.F.(2005). Biomarkers, health, lifestyle, and demographic variables as correlates of reaction time performance in early, middle, and late adulthood. Q J Exp Psychol A 58, 5-21.
Barraclough, N.E., and Perrett, D.I.(2011). From single cells to social perception. Philosophical Transactions of the Royal Society B-Biological Sciences 366, 1739-1752.
Bate, S., Adams, A., Bennetts, R., and Line, H.(2019). Developmental prosopagnosia with concurrent topographical difficulties:A case report and virtual reality training programme. Neuropsychol Rehabil 29, 1290-1312.
Bate, S., Cook, S.J., Duchaine, B., Tree, J.J., Burns, E.J., and Hodgson, T.L.(2014). Intranasal inhalation of oxytocin improves face processing in developmental prosopagnosia. Cortex 50, 55-63.
Baylis, G.C., Rolls, E.T., and Leonard, C.M.(1985). Selectivity between Faces in the Responses of a Population of Neurons in the Cortex in the Superior Temporal Sulcus of the Monkey. Brain Res 342, 91-102.
Behrmann, M., and Avidan, G.(2005). Congenital prosopagnosia:face-blind from birth. Trends Cogn Sci 9, 180-187.
Bentin, S., Degutis, J.M., D'Esposito, M., and Robertson, L.C.(2007). Too many trees to see the forest:performance, event-related potential, and functional magnetic resonance imaging manifestations of integrative congenital prosopagnosia. J Cogn Neurosci 19, 132-146.
Bodamer, J.(1947). Die Prosop-Agnosie. Archiv für Psychiatrie und Nervenkrankheiten. 179, 6-53.
Bolger, A.M., Lohse, M., and Usadel, B.(2014). Trimmomatic:a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
Bowles, D.C., McKone, E., Dawel, A., Duchaine, B., Palermo, R., Schmalzl, L., Rivolta, D., Wilson, C.E., and Yovel, G.(2009). Diagnosing prosopagnosia:effects of ageing, sex, and participant-stimulus ethnic match on the Cambridge Face Memory Test and Cambridge Face Perception Test. Cogn Neuropsychol 26, 423-455.
Bruce, C., Desimone, R., and Gross, C.G.(1981). Visual Properties of Neurons in a Polysensory Area in Superior Temporal Sulcus of the Macaque. Journal of Neurophysiology 46, 369-384.
Bruce, V., and Young, A.(1986). Understanding face recognition. Br J Psychol 77( Pt 3), 305-327.
Burianova, H., Lee, Y., Grady, C.L., and Moscovitch, M.(2013). Age-related dedifferentiation and compensatory changes in the functional network underlying face processing. Neurobiol Agin g34, 2759-2767.
Busigny, T., and Rossion, B.(2010). Acquired prosopagnosia abolishes the face inversion effect. Cortex 46, 965-981.
Carbon, C.C., Gruter, T., Gruter, M., Weber, J.E., and Lueschow, A.(2010). Dissociation of facial attractiveness and distinctiveness processing in congenital prosopagnosia. Vis Cogn 18, 641-654.
Carbon, C.C., Gruter, T., Weber, J.E., and Lueschow, A.(2007). Faces as objects of non-expertise:processing of thatcherised faces in congenital prosopagnosia. Perception 36, 1635-1645.
Chen, B.Q., Zhu, Z.J., Na, R., Fang, W., Zhang, W.X., Zhou, Q., Zhou, S.B., Lei, H., Huang, A.L., Chen, T.M., et al.(2018a). Genomic Analyses of Visual Cognition:Perceptual Rivalry and Top-Down Control. Journal of Neuroscience 38, 9668-9678.
Chen, B.Q., Zhu, Z.J., Wang, Y.Y., Ding, X.H., Guo, X.B., He, M.G., Fang, W., Zhou, Q., Zhou, S.B., Lei, H., et al.(2018b). Nature vs. nurture in human sociality:multi-level genomic analyses of social conformity. Journal of Human Genetics 63, 605-619.
Chiang, C., Layer, R.M., Faust, G.G., Lindberg, M.R., Rose, D.B., Garrison, E.P., Marth, G.T., Quinlan, A.R., and Hall, I.M.(2015). SpeedSeq:ultra-fast personal genome analysis and interpretation. Nat Methods 12, 966-968.
Chitty, A.J., Perrett, D.I., Mistlin, A.J., and Potter, D.D.(1985). Visual Cells in the Temporal Cortex Selectively Responsive to the Sight of Hands Manipulating Objects. Perception 14, A15-A15.
Cho, W., and Stahelin, R.V.(2006). Membrane binding and subcellular targeting of C2 domains. Biochimica et Biophysica Acta(BBA)- Molecular and Cell Biology of Lipids1761, 838-849.
Cirulli, E.T., and Goldstein, D.B.(2010). Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 11, 415-425.
Clark, V.P., Keil, K., Maisog, J.M., Courtney, S., Ungerleider, L.G., and Haxby, J.V.(1996). Functional magnetic resonance imaging of human visual cortex during face matching:A comparison with positron emission tomography. Neuroimage 4, 1-15.
Cong, P.K., Bai, W.Y., Li, J.C., Yang, M.Y., Khederzadeh, S., Gai, S.R., Li, N., Liu, Y.H., Yu, S.H., Zhao, W.W., et al.(2022a). Genomic analyses of 10,376 individuals in the Westlake BioBank for Chinese(WBBC)pilot project. Nat Commun 13, 2939.
Cong, P.K., Khederzadeh, S., Yuan, C.D., Ma, R.J., Zhang, Y.Y., Liu, J.Q., Yu, S.H., Xu, L., Gao, J.H., Pan, H.X., et al.(2022b). Identification of clinically actionable secondary genetic variants from whole-genome sequencing in a large-scale Chinese population. Clin Transl Med 12, e866.
Crawford, J.R., and Garthwaite, P.H.(2002). Investigation of the single case in neuropsychology:confidence limits on the abnormality of test scores and test score differences. Neuropsychologia 40, 1196-1208.
Crawford, J.R., Garthwaite, P.H., and Porter, S.(2010). Point and interval estimates of effect sizes for the case-controls design in neuropsychology:rationale, methods, implementations, and proposed reporting standards. Cogn Neuropsychol 27, 245-260.
Crawford, J.R., and Howell, D.C.(1998). Comparing an individual's test score against norms derived from small samples. Clinical Neuropsychologist 12, 482-486.
Dalrymple, K.A., and Palermo, R.(2016). Guidelines for studying developmental prosopagnosia in adults and children. Wiley Interdiscip Rev Cogn Sci 7, 73-87.
Damasio, A.R., Tranel, D., and Damasio, H.(1990). Face agnosia and the neural substrates of memory. Annu Rev Neurosci 13, 89-109.
De Haan, E.H.(1999). A familial factor in the development of face recognition deficits. J Clin Exp Neuropsychol21, 312-315.
Dennett, H.W., McKone, E., Tavashmi, R., Hall, A., Pidcock, M., Edwards, M., and Duchaine, B.(2012). The Cambridge Car Memory Test:a task matched in format to the Cambridge Face Memory Test, with norms, reliability, sex differences, dissociations from face memory, and expertise effects. Behav Res Methods 44, 587-605.
Desimone, R., Albright, T.D., Gross, C.G., and Bruce, C.(1984). Stimulus-Selective Properties of Inferior Temporal Neurons in the Macaque. Journal of Neuroscience 4, 2051-2062.
Dinkelacker, V., Gruter, M., Klaver, P., Gruter, T., Specht, K., Weis, S., Kennerknecht, I., Elger, C.E., and Fernandez, G.(2011). Congenital prosopagnosia:multistage anatomical and functional deficits in face processing circuitry. J Neurol 258, 770-782.
Djurovic, S., Le Hellard, S., Kahler, A.K., Jonsson, E.G., Agartz, I., Steen, V.M., Hall, H., Wang, A.G., Rasmussen, H.B., Melle, I., et al.(2009). Association of MCTP2 gene variants with schizophrenia in three independent samples of Scandinavian origin(SCOPE). Psychiatry Res 168, 256-258.
Dobel, C., Bolte, J., Aicher, M., and Schweinberger, S.R.(2007). Prosopagnosia without apparent cause:overview and diagnosis of six cases. Cortex 43, 718-733.
Dricot, L., Sorger, B., Schiltz, C., Goebel, R., and Rossion, B.(2008). The roles of “face” and “non-face” areas during individual face perception:Evidence by fMRI adaptation in a brain-damaged prosopagnosic patient. NeuroImage 40, 318-332.
Duchaine, B., Germine, L., and Nakayama, K.(2007). Family resemblance:ten family members with prosopagnosia and within-class object agnosia. Cogn Neuropsychol 24, 419-430.
Duchaine, B., and Nakayama, K.(2006a). The Cambridge Face Memory Test:results for neurologically intact individuals and an investigation of its validity using inverted face stimuli and prosopagnosic participants. Neuropsychologia 44, 576-585.
Duchaine, B.C., and Nakayama, K.(2004). Developmental prosopagnosia and the Benton Facial Recognition Test. Neurology 62, 1219-1220.
Duchaine, B.C., and Nakayama, K.(2006b). Developmental prosopagnosia:a window to content-specific face processing. Curr Opin Neurobiol 16, 166-173.
Duchaine, B.C., and Weidenfeld, A.(2003). An evaluation of two commonly used tests of unfamiliar face recognition. Neuropsychologia 41, 713-720.
Dunning, D., Johnson, K., Ehrlinger, J., and Kruger, J.(2003). Why People Fail to Recognize Their Own Incompetence. 12, 83-87.
Dzhelyova, M.P., Ellison, A., and Atkinson, A.P.(2011). Event-related repetitive TMS reveals distinct, critical roles for right OFA and bilateral posterior STS in judging the sex and trustworthiness of faces. J Cogn Neurosci 23, 2782-2796.
Esins, J., Schultz, J., Stemper, C., Kennerknecht, I., and Bulthoff, I.(2016). Face Perception and Test Reliabilities in Congenital Prosopagnosia in Seven Tests. Iperception 7, 2041669515625797.
Esins, J., Schultz, J., Stemper, C., Kennerknecht, I., Wallraven, C., and Bulthoff, I.(2015). Corrigendum:Do congenital prosopagnosia and the other-race effect affect the same face recognition mechanisms? Front Hum Neurosci 9, 294.
Espino-Saldaña, A.E., Durán-Ríos, K., Olivares-Hernandez, E., Rodríguez-Ortiz, R., Arellano-Carbajal, F., and Martínez-Torres, A.(2020). Temporal and spatial expression of zebrafish mctp genes and evaluation of frameshift alleles of mctp2b. Gene 738, 144371.
Forshew, T., Murtaza, M., Parkinson, C., Gale, D., Tsui, D.W., Kaper, F., Dawson, S.J., Piskorz, A.M., Jimenez-Linan, M., Bentley, D., et al.(2012). Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 4, 136ra168.
Freiwald, W., Duchaine, B., and Yovel, G.(2016). Face Processing Systems:From Neurons to Real-World Social Perception. Annu Rev Neurosci 39, 325-346.
Galaburda, A.M., and Duchaine, B.C.(2003). Developmental disorders of vision. Neurol Clin 21, 687-707.
Gao, X., Vuong, Q.C., and Rossion, B.(2019). The cortical face network of the prosopagnosic patient PS with fast periodic stimulation in fMRI. Cortex 119, 528-542.
Genc, O., Dickman, D.K., Ma, W., Tong, A., Fetter, R.D., and Davis, G.W.(2017). MCTP is an ER-resident calcium sensor that stabilizes synaptic transmission and homeostatic plasticity. Elife6.
Genomes Project, C., Abecasis, G.R., Altshuler, D., Auton, A., Brooks, L.D., Durbin, R.M., Gibbs, R.A., Hurles, M.E., and McVean, G.A.(2010). A map of human genome variation from population-scale sequencing. Nature4 67, 1061-1073.
Genomes Project, C., Abecasis, G.R., Auton, A., Brooks, L.D., DePristo, M.A., Durbin, R.M., Handsaker, R.E., Kang, H.M., Marth, G.T., and McVean, G.A.(2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56-65.
Genomes Project, C., Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Korbel, J.O., Marchini, J.L., McCarthy, S., McVean, G.A., et al.(2015). A global reference for human genetic variation. Nature 526, 68-74.
Germine, L.T., Duchaine, B., and Nakayama, K.(2011). Where cognitive development and aging meet:face learning ability peaks after age 30. Cognition 118, 201-210.
Goh, J.O., Suzuki, A., and Park, D.C.(2010). Reduced neural selectivity increases fMRI adaptation with age during face discrimination. Neuroimage 51, 336-344.
Grill-Spector, K., Henson, R., and Martin, A.(2006). Repetition and the brain:neural models of stimulus-specific effects. Trends Cogn Sci 10, 14-23.
Grill-Spector, K., Knouf, N., and Kanwisher, N.(2004). The fusiform face area subserves face perception, not generic within-category identification. Nat Neurosci 7, 555-562.
Grill-Spector, K., and Malach, R.(2001). fMR-adaptation:a tool for studying the functional properties of human cortical neurons. Acta Psychol(Amst)107, 293-321.
Grill-Spector, K., and Malach, R.(2004). The human visual cortex. Annu Rev Neurosci 27, 649-677.
Gross, C.G., Rochamir.Ce, and Bender, D.B.(1972). Visual Properties of Neurons in Inferotemporal Cortex of Macaque. Journal of Neurophysiology 35, 96-&.
Grueter, M., Grueter, T., Bell, V., Horst, J., Laskowski, W., Sperling, K., Halligan, P.W., Ellis, H.D., and Kennerknecht, I.(2007). Hereditary prosopagnosia:the first case series. Cortex 43, 734-749.
Gruter, T., Gruter, M., Bell, V., and Carbon, C.C.(2009). Visual mental imagery in congenital prosopagnosia. Neurosci Lett 453, 135-140.
Gruter, T., Gruter, M., and Carbon, C.C.(2008). Neural and genetic foundations of face recognition and prosopagnosia. J Neuropsychol 2, 79-97.
Gusella, J.F., Wexler, N.S., Conneally, P.M., Naylor, S.L., Anderson, M.A., Tanzi, R.E., Watkins, P.C., Ottina, K., Wallace, M.R., Sakaguchi, A.Y., et al.(1983). A polymorphic DNA marker genetically linked to Huntington's disease. Nature 306, 234-238.
Hadjikhani, N., and de Gelder, B.(2002). Neural basis of prosopagnosia:an fMRI study. Hum Brain Mapp 16, 176-182.
Haxby, J.V., Hoffman, E.A., and Gobbini, M.I.(2000). The distributed human neural system for face perception. Trends Cogn Sci 4, 223-233.
Haxby, J.V., Horwitz, B., Ungerleider, L.G., Maisog, J.M., Pietrini, P., and Grady, C.L.(1994). The Functional-Organization of Human Extrastriate Cortex - a Pet-Rcbf Study of Selective Attention to Faces and Locations. Journal of Neuroscience 14, 6336-6353.
Heinzen, E.L., Neale, B.M., Traynelis, S.F., Allen, A.S., and Goldstein, D.B.(2015). The genetics of neuropsychiatric diseases:looking in and beyond the exome. Annu Rev Neurosci 38, 47-68.
Henson, R.N., and Rugg, M.D.(2003). Neural response suppression, haemodynamic repetition effects, and behavioural priming. Neuropsychologia 41, 263-270.
Hung, C.C., Yen, C.C., Ciuchta, J.L., Papoti, D., Bock, N.A., Leopold, D.A., and Silva, A.C.(2015). Functional mapping of face-selective regions in the extrastriate visual cortex of the marmoset. J Neurosci 35, 1160-1172.
Johnen, A., Schmukle, S.C., Huttenbrink, J., Kischka, C., Kennerknecht, I., and Dobel, C.(2014). A family at risk:congenital prosopagnosia, poor face recognition and visuoperceptual deficits within one family. Neuropsychologia 58, 52-63.
Jordan, D.M., Ramensky, V.E., and Sunyaev, S.R.(2010). Human allelic variation:perspective from protein function, structure, and evolution. Curr Opin Struct Biol 20, 342-350.
Kanwisher, N., McDermott, J., and Chun, M.M.(1997). The fusiform face area:a module in human extrastriate cortex specialized for face perception. J Neurosci 17, 4302-4311.
Kennerknecht, I.(2021). Prevalence of Hereditary Prosopagnosia – A Worldwide Survey. . Münster:Westfälische Wilhelms-Universität Münster
Kennerknecht, I., Grueter, T., Welling, B., Wentzek, S., Horst, J., Edwards, S., and Grueter, M.(2006). First report of prevalence of non-syndromic hereditary prosopagnosia(HPA). Am J Med Genet A 140, 1617-1622.
Kennerknecht, I., Ho, N.Y., and Wong, V.C.(2008a). Prevalence of hereditary prosopagnosia(HPA)in Hong Kong Chinese population. Am J Med Genet A 146A, 2863-2870.
Kennerknecht, I., Pluempe, N., and Welling, B.(2008b). Congenital prosopagnosia--a common hereditary cognitive dysfunction in humans. Front Biosci 13, 3150-3158.
Kennerknecht, I., Plumpe, N., Edwards, S., and Raman, R.(2007). Hereditary prosopagnosia(HPA):the first report outside the Caucasian population. Journal of Human Genetics 52, 230-236.
Kreiman, G., Koch, C., and Fried, I.(2000). Category-specific visual responses of single neurons in the human medial temporal lobe. Nat Neurosci 3, 946-953.
Kress, T., and Daum, I.(2003). Developmental prosopagnosia:a review. Behav Neurol14, 109-121.
Kruger, J., and Dunning, D.(1999). Unskilled and unaware of it:how difficulties in recognizing one's own incompetence lead to inflated self-assessments. J Pers Soc Psychol 77, 1121-1134.
Kweon, K., Shin, E.S., Park, K.J., Lee, J.K., Joo, Y., and Kim, H.W.(2018). Genome-Wide Analysis Reveals Four Novel Loci for Attention-Deficit Hyperactivity Disorder in Korean Youths. Soa--ch'ongsonyon chongsin uihak = Journal of child & adolescent psychiatry 29, 62-72.
Le Grand, R., Cooper, P.A., Mondloch, C.J., Lewis, T.L., Sagiv, N., de Gelder, B., and Maurer, D.(2006). What aspects of face processing are impaired in developmental prosopagnosia? Brain Cogn 61, 139-158.
Lee, S., Wu, M.C., and Lin, X.(2012). Optimal tests for rare variant effects in sequencing association studies. Biostatistics 13, 762-775.
Lee, Y., Duchaine, B., Wilson, H.R., and Nakayama, K.(2010). Three cases of developmental prosopagnosia from one family:detailed neuropsychological and psychophysical investigation of face processing. Cortex 46, 949-964.
Lee, Y., Grady, C.L., Habak, C., Wilson, H.R., and Moscovitch, M.(2011). Face processing changes in normal aging revealed by fMRI adaptation. J Cogn Neurosci 23, 3433-3447.
Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O'Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B., et al.(2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285-291.
Li, H., and Durbin, R.(2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760.
Liu, J., Harris, A., and Kanwisher, N.(2010). Perception of Face Parts and Face Configurations:An fMRI Study. Journal of Cognitive Neuroscience 22, 203-211.
Malach, R., Reppas, J.B., Benson, R.R., Kwong, K.K., Jiang, H., Kennedy, W.A., Ledden, P.J., Brady, T.J., Rosen, B.R., and Tootell, R.B.H.(1995). Object-Related Activity Revealed by Functional Magnetic-Resonance-Imaging in Human Occipital Cortex. P Natl Acad Sci USA 92, 8135-8139.
Manichaikul, A., Mychaleckyj, J.C., Rich, S.S., Daly, K., Sale, M., and Chen, W.M.(2010). Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867-2873.
McCall, M.N., Uppal, K., Jaffee, H.A., Zilliox, M.J., and Irizarry, R.A.(2011). The Gene Expression Barcode:leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res 39, D1011-1015.
McCarthy, G., Puce, A., Belger, A., and Allison, T.(1999). Electrophysiological studies of human face perception. II:Response properties of face-specific potentials generated in occipitotemporal cortex. Cereb Cortex 9, 431-444.
McCarthy, G., Puce, A., Gore, J.C., and Allison, T.(1997). Face-specific processing in the human fusiform gyrus. J Cogn Neurosci 9, 605-610.
McConachie, H.R.(1976). Developmental prosopagnosia. A single case report. Cortex 12, 76-82.
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., et al.(2010). The Genome Analysis Toolkit:a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297-1303.
McKone, E., and Palermo, R.(2010). A strong role for nature in face recognition. Proc Natl Acad Sci U S A 107, 4795-4796.
McKone, E., Stokes, S., Liu, J., Cohan, S., Fiorentini, C., Pidcock, M., Yovel, G., Broughton, M., and Pelleg, M.(2012). A robust method of measuring other-race and other-ethnicity effects:the Cambridge Face Memory Test format. PLoS One 7, e47956.
Mick, E., Todorov, A., Smalley, S., Hu, X., Loo, S., Todd, R.D., Biederman, J., Byrne, D., Dechairo, B., Guiney, A., et al.(2010). Family-Based Genome-Wide Association Scan of Attention-Deficit/Hyperactivity Disorder. Journal of the American Academy of Child & Adolescent Psychiatry 49, 898-905.e893.
Moeller, S., Crapse, T., Chang, L., and Tsao, D.Y.(2017). The effect of face patch microstimulation on perception of faces and objects. Nat Neurosci 20, 743-+.
Moeller, S., Freiwald, W.A., and Tsao, D.Y.(2008). Patches with links:a unified system for processing faces in the macaque temporal lobe. Science 320, 1355-1359.
Ng, P.C., and Henikoff, S.(2001). Predicting deleterious amino acid substitutions. Genome Res 11, 863-874.
Nunn, J.A., Postma, P., and Pearson, R.(2001). Developmental prosopagnosia:should it be taken at face value? Neurocase 7, 15-27.
Park, J., Carp, J., Kennedy, K.M., Rodrigue, K.M., Bischof, G.N., Huang, C.M., Rieck, J.R., Polk, T.A., and Park, D.C.(2012). Neural broadening or neural attenuation? Investigating age-related dedifferentiation in the face network in a large lifespan sample. J Neurosci 32, 2154-2158.
Perrett, D.I., Mistlin, A.J., and Chitty, A.J.(1987). Visual Neurons Responsive to Faces. Trends in Neurosciences 10, 358-364.
Perrett, D.I., Rolls, E.T., and Caan, W.(1982). Visual Neurones Responsive to Faces in the Monkey Temporal Cortex. Exp Brain Res 47, 329-342.
Perrett, D.I., Smith, P.A.J., Potter, D.D., Mistlin, A.J., Head, A.S., Milner, A.D., and Jeeves, M.A.(1984). Neurons Responsive to Faces in the Temporal Cortex - Studies of Functional-Organization, Sensitivity to Identity and Relation to Perception. Human Neurobiology 3, 197-208.
Perrett, D.I., Smith, P.A.J., Potter, D.D., Mistlin, A.J., Head, A.S., Milner, A.D., and Jeeves, M.A.(1985). Visual Cells in the Temporal Cortex Sensitive to Face View and Gaze Direction. Proceedings of the Royal Society Series B-Biological Sciences 223, 293-317.
Pitcher, D., Charles, L., Devlin, J.T., Walsh, V., and Duchaine, B.(2009). Triple Dissociation of Faces, Bodies, and Objects in Extrastriate Cortex. Curr Biol 19, 319-324.
Pitcher, D., Garrido, L., Walsh, V., and Duchaine, B.C.(2008). Transcranial magnetic stimulation disrupts the perception and embodiment of facial expressions. J Neurosci 28, 8929-8933.
Poelmans, G., Pauls, D.L., Buitelaar, J.K., and Franke, B.(2011). Integrated genome-wide association study findings:identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiat 168, 365-377.
Polk, T.A., Park, J., Smith, M.R., and Park, D.C.(2007). Nature versus nurture in ventral visual cortex:A functional magnetic resonance Imaging study of twins. J Neurosci 27, 13921-13925.
Pritchard, J.K.(2001). Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet 69, 124-137.
Puce, A., Allison, T., Asgari, M., Gore, J.C., and McCarthy, G.(1996). Differential Sensitivity of Human Visual Cortex to Faces, Letterstrings, and Textures:A Functional Magnetic Resonance Imaging Study. J Neurosci 16, 5205-5215.
Puce, A., Allison, T., and McCarthy, G.(1999). Electrophysiological studies of human face perception. III:Effects of top-down processing on face-specific potentials. Cereb Cortex 9, 445-458.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A., Bender, D., Maller, J., Sklar, P., de Bakker, P.I., Daly, M.J., et al.(2007). PLINK:a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559-575.
Purcell, S.M., Moran, J.L., Fromer, M., Ruderfer, D., Solovieff, N., Roussos, P., O'Dushlaine, C., Chambert, K., Bergen, S.E., Kahler, A., et al.(2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185-190.
Qiu, L., Yu, H., and Liang, F.(2015). Multiple C2 domains transmembrane protein 1 is expressed in CNS neurons and possibly regulates cellular vesicle retrieval and oxidative stress. J Neurochem 135, 492-507.
Rangarajan, V., Hermes, D., Foster, B.L., Weiner, K.S., Jacques, C., Grill-Spector, K., and Parvizi, J.(2014). Electrical stimulation of the left and right human fusiform gyrus causes different effects in conscious face perception. J Neurosci 34, 12828-12836.
Rasetti, R., and Weinberger, D.R.(2011). Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev 21, 340-348.
Righart, R., Andersson, F., Schwartz, S., Mayer, E., and Vuilleumier, P.(2009). Top-Down Activation of Fusiform Cortex without Seeing Faces in Prosopagnosia. Cerebral Cortex 20, 1878-1890.
Robinson, J.T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E.S., Getz, G., and Mesirov, J.P.(2011). Integrative genomics viewer. Nat Biotechnol 29, 24-26.
Rolls, E.T.(1984). Neurons in the Cortex of the Temporal-Lobe and in the Amygdala of the Monkey with Responses Selective for Faces. Hum Neurobiol 3, 209-222.
Rolls, E.T., Baylis, G.C., and Leonard, C.M.(1985). Role of Low and High Spatial-Frequencies in the Face-Selective Responses of Neurons in the Cortex in the Superior Temporal Sulcus in the Monkey. Vision Res 25, 1021-1035.
Rossion, B., Dricot, L., Goebel, R., and Busigny, T.(2011). Holistic Face Categorization in Higher Order Visual Areas of the Normal and Prosopagnosic Brain:Toward a Non-Hierarchical View of Face Perception. 4.
Rotshtein, P., Henson, R.N., Treves, A., Driver, J., and Dolan, R.J.(2005). Morphing Marilyn into Maggie dissociates physical and identity face representations in the brain. Nat Neurosci 8, 107-113.
Saito, H., Yukie, M., Tanaka, K., Hikosaka, K., Fukada, Y., and Iwai, E.(1986). Integration of Direction Signals of Image Motion in the Superior Temporal Sulcus of the Macaque Monkey. Journal of Neuroscience 6, 145-157.
Schiltz, C., and Rossion, B.(2006). Faces are represented holistically in the human occipito-temporal cortex. Neuroimage 32, 1385-1394.
Schiltz, C., Sorger, B., Caldara, R., Ahmed, F., Mayer, E., Goebel, R., and Rossion, B.(2006). Impaired face discrimination in acquired prosopagnosia is associated with abnormal response to individual faces in the right middle fusiform gyrus. Cereb Cortex 16, 574-586.
Schmalzl, L., Palermo, R., and Coltheart, M.(2008). Cognitive heterogeneity in genetically based prosopagnosia:a family study. J Neuropsycho l2, 99-117.
Schrott, H.G., Goldstein, J.L., Hazzard, W.R., McGoodwin, M.M., and Motulsky, A.G.(1972). Familial hypercholesterolemia in a large indred. Evidence for a monogenic mechanism. Ann Intern Med 76, 711-720.
Scott, L.J., Muglia, P., Kong, X.Q., Guan, W., Flickinger, M., Upmanyu, R., Tozzi, F., Li, J.Z., Burmeister, M., Absher, D., et al.(2009). Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A 106, 7501-7506.
Sergent, J., Ohta, S., and Macdonald, B.(1992). Functional Neuroanatomy of Face and Object Processing - a Positron Emission Tomography Study. Brain115, 15-36.
Shah, P., Gaule, A., Sowden, S., Bird, G., and Cook, R.(2015). The 20-item prosopagnosia index(PI20):a self-report instrument for identifying developmental prosopagnosia. R Soc Open Sci 2, 140343.
Sherry, S.T., Ward, M.H., Kholodov, M., Baker, J., Phan, L., Smigielski, E.M., and Sirotkin, K.(2001). dbSNP:the NCBI database of genetic variation. Nucleic Acids Res 29, 308-311.
Shin, O.H., Han, W., Wang, Y., and Sudhof, T.C.(2005). Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions(MCTPs)and unusual Ca2+ binding properties. J Biol Chem 280, 1641-1651.
Shupliakov, O., and Brodin, L.(2010). Recent insights into the building and cycling of synaptic vesicles. Exp Cell Res 316, 1344-1350.
Sjostedt, E., Zhong, W., Fagerberg, L., Karlsson, M., Mitsios, N., Adori, C., Oksvold, P., Edfors, F., Limiszewska, A., Hikmet, F., et al.(2020). An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367.
Stollhoff, R., Jost, J., Elze, T., and Kennerknecht, I.(2010). The early time course of compensatory face processing in congenital prosopagnosia. PLoS One 5, e11482.
Stollhoff, R., Jost, J., Elze, T., and Kennerknecht, I.(2011). Deficits in long-term recognition memory reveal dissociated subtypes in congenital prosopagnosia. PLoS One 6, e15702.
Susilo, T., and Duchaine, B.(2013). Advances in developmental prosopagnosia research. Current Opinion in Neurobiology 23, 423-429.
Thorvaldsdottir, H., Robinson, J.T., and Mesirov, J.P.(2013). Integrative Genomics Viewer(IGV):high-performance genomics data visualization and exploration. Brief Bioinform 14, 178-192.
Tsao, D.Y., Freiwald, W.A., Knutsen, T.A., Mandeville, J.B., and Tootell, R.B.(2003). Faces and objects in macaque cerebral cortex. Nat Neurosci 6, 989-995.
Tsao, D.Y., Freiwald, W.A., Tootell, R.B., and Livingstone, M.S.(2006). A cortical region consisting entirely of face-selective cells. Science 311, 670-674.
Tsao, D.Y., and Livingstone, M.S.(2008). Mechanisms of face perception. Annu Rev Neurosci 31, 411-437.
Tsao, D.Y., Moeller, S., and Freiwald, W.A.(2008). Comparing face patch systems in macaques and humans. Proc Natl Acad Sci U S A 105, 19514-19519.
Tsui, L.C., Buchwald, M., Barker, D., Braman, J.C., Knowlton, R., Schumm, J.W., Eiberg, H., Mohr, J., Kennedy, D., Plavsic, N., et al.(1985). Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science 230, 1054-1057.
Verfaillie, K., Huysegems, S., De Graef, P., and Van Belle, G.(2014). Impaired holistic and analytic face processing in congenital prosopagnosia:Evidence from the eye-contingent mask/window paradigm. Vis Cogn 22, 503-521.
von Kriegstein, K., Dogan, O., Gruter, M., Giraud, A.L., Kell, C.A., Gruter, T., Kleinschmidt, A., and Kiebel, S.J.(2008). Simulation of talking faces in the human brain improves auditory speech recognition. Proc Natl Acad Sci U S A 105, 6747-6752.
Wang, K., Li, M., Hadley, D., Liu, R., Glessner, J., Grant, S.F., Hakonarson, H., and Bucan, M.(2007). PennCNV:an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17, 1665-1674.
Wang, K., Li, M., and Hakonarson, H.(2010). ANNOVAR:functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164.
White, D., and Burton, A.M.(2022). Individual differences and the multidimensional nature of face perception. Nature Reviews Psychology 1, 287-300.
Wilmer, J.B., Germine, L., Chabris, C.F., Chatterjee, G., Williams, M., Loken, E., Nakayama, K., and Duchaine, B.(2010). Human face recognition ability is specific and highly heritable. P Natl Acad Sci USA 107, 5238-5241.
Yamane, S., Kaji, S., and Kawano, K.(1988). What Facial Features Activate Face Neurons in the Inferotemporal Cortex of the Monkey. Exp Brain Res 73, 209-214.
Yovel, G., and Kanwisher, N.(2004). Face perception:domain specific, not process specific. Neuron 44, 889-898.
Yovel, G., and Kanwisher, N.(2005). The neural basis of the behavioral face-inversion effect. Curr Biol 15, 2256-2262.
Zebrowitz, L., Ward, N., Boshyan, J., Gutchess, A., and Hadjikhani, N.(2016). Dedifferentiated face processing in older adults is linked to lower resting state metabolic activity in fusiform face area. Brain Res 1644, 22-31.
Zhang, J., Li, X., Song, Y., and Liu, J.(2012). The fusiform face area is engaged in holistic, not parts-based, representation of faces. PLoS One7, e40390.
Zhao, Y., Zhen, Z., Liu, X., Song, Y., and Liu, J.(2018). The neural network for face recognition:Insights from an fMRI study on developmental prosopagnosia. Neuroimage 169, 151-161.
Zhao, Y.F., Li, J.G., Liu, X.Q., Song, Y.Y., Wang, R.S., Yang, Z.T., and Liu, J.(2016). Altered spontaneous neural activity in the occipital face area reflects behavioral deficits in developmental prosopagnosia. Neuropsychologia 89, 344-355.
Zhou, X., and Jenkins, R.(2020). Dunning-Kruger effects in face perception. Cognition 203, 104345.
Zhu, Q., Song, Y.Y., Hu, S.Y., Li, X.B., Tian, M.Q., Zhen, Z.L., Dong, Q., Kanwisher, N., and Liu, J.(2010). Heritability of the Specific Cognitive Ability of Face Perception. Current Biology 20, 137-142.
Zhu, X.W., Liu, K.Q., Wang, P.Y., Liu, J.Q., Chen, J.Y., Xu, X.J., Xu, J.J., Qiu, M.C., Sun, Y., Liu, C., et al.(2021). Cohort profile:the Westlake BioBank for Chinese(WBBC)pilot project. BMJ Open 11, e045564.
Zhu, Z., Chen, B., Na, R., Fang, W., Zhang, W., Zhou, Q., Zhou, S., Lei, H., Huang, A., Chen, T., et al.(2019). Heritability of human visual contour integration-an integrated genomic study. Eur J Hum Genet 27, 1867-1875.
Zhu, Z., Wang, Y., Cao, Z., Chen, B., Cai, H., Wu, Y., and Rao, Y.(2016). Cue-independent memory impairment by reactivation-coupled interference in human declarative memory. Cognition 155, 125-134.
Zhu, Z.J., Chen, B.Q., Yan, H.M., Fang, W., Zhou, Q., Zhou, S.B., Lei, H., Huang, A.L., Chen, T.M., Gao, T.M., et al.(2018). Multi-level genomic analyses suggest new genetic variants involved in human memory. Eur J Hum Genet 26, 1668-1678.
Zhu, Z., Chen, B.Q., Na, R., Fang, W., Zhang, W.X., Zhou, Q., Zhou, S.B., Lei, H., Huang, A.L., Chen, T.M., Gao, T.M., Chen, L., Ni, D.S., Gu., Y.P., Liu, J.N., Chen, J.Y., Rao, Y. and Fang, F.(2020). Heritability of human visual controur integration--an integrated genomic study. Eur J Hum Genet 27, 1867-1875.
图例

图1:Mapping of a Novel Autosomal Dominant CP Locus to the Short Arm of Chromosome 15(15q26).(a)The pedigree plot of family A with genotypes for c.2147T>G(p.I716S), GenBank:NM_018349 and phenotypes. Crosses, DNA samples not available; diagonal slashes, dead members; filled symbols, family members poor on both interviews and the behavioral CFMT test ; half-filled symbols, family members who had daily face recognition problems but appeared normal on CFMT or founders with daily face recognition problems ; an arrow head, the index of family A.
(b)Graphical representation of parametric linkage results of linkage 1 on chr15(LOD score = 3.49)in family A with diagnosis based only on interviews as well as the CFMT behavioral test, assuming a rare dominant model.
(c)Graphical representation of parametric linkage results of linkage 2 on chr15(LOD score = 5.13)in family A with diagnosis based on interviews not behavioral tests, assuming a rare dominant model.

图2:Mutations Detection in MCTP2 and Co-segregation in CP.
(a)The distribution of the 20-item questionnaire scores in 743 males(the blue approximate normal curve)and 2161 females(the red approximate normal curve). Dashed lines indicate the cut-off of 2 SDs poorer than the mean. Sticks with different colors indicate different individuals with different mutations.
(b)A mutation c.239delG, leading to p.S80fs in individuals B0001, B0009, B0046 with CP. All three of them were aware of difficulties with face recognition early in their life(Table S2).
(c)Correlation of the(c.239delG, p.S80fs)genotype with the CP phenotype in the family of B0001, with 3 members tested.
(d)A mutation c.814A>T, leading to p. M272L in individual B0011.
(e)Correlation of the genotype(c.814A>T [p.M272L])with the CP phenotype in the family of B0011.
(f)A mutation c.1120A>G, leading to p.T374A in individual B0010.
(g)A mutation c.1642G>A, leading to p.V548I in individual B0003.
(h)Correlation of the(c.1642G>A [p.V548I])genotype with the CP phenotype in the family of B0003, with 6 members tested(Table S2).
(i)A mutation c.1922G>A, leading to p.R641Q in individual B0002.
(j)Correlation of the(c.1922G>A [p.R641Q])genotype with the CP phenotype in the family of B0002, with 5 members tested(Table S2).

图3:Co-segregation of c.239delG(p.S80fs)in MCTP2 in CP.
16 individuals with the frameshift deletion mutation(c.239delG [p.S80fs])were detected in a cohort of 1757 students. 14 of them were available for further analysis. More members from 6 families were available(Table S3).
(a)Correlation of the(c.239delG [p.S80fs])genotype with the CP phenotype in the family of C2666, with 10 family members tested.
(b)Correlation of the(c.239delG [p.S80fs])genotype with the CP phenotype in the family of C3164, with 4 family members tested.
(c)Correlation of the(c.239delG [p.S80fs])genotype with the CP phenotype in the family of C3049 with 6 family members tested.
(d)Correlation of the(c.239delG [p.S80fs])genotype with the CP phenotype in the family of C2149 with 4 family members tested.
(e)Correlation of the(c.239delG [p.S80fs])genotype with the CP phenotype in the family of C3030, with 3 members tested.
(f)The pedigree plot of family C3649 with genotype and phenotype.

图4:rFFA Activation in Face Discrimination
Experimental paradigm as Figure 4a. The average percent signal change(±SD for controls)from baseline fixation is plotted for the different and identical upright face conditions. Bold response to pairs of upright different faces or identical faces in the rFFA of controls(n=20)(b); non-CP members A-VI:1(c), A-VI:2(d), A-VI:10(e)and CP members A-VI:5(f), A-VI:7(g), and A-VI:9(h)from family A with the I716S mutation inMCTP2; non-CP C2666-III:2(i)and CP C2666-III:4(j)from family C2666 with the S80fs mutation in MCTP2.

图5:Comparison between CP and Control Individuals in the Adaptation Experiment
The adaptation score(difference in percent signal change between responses to upright different faces and upright identical faces)is plotted for CP individuals(black bars)and each individual control subject(white bars)in an increasing order.
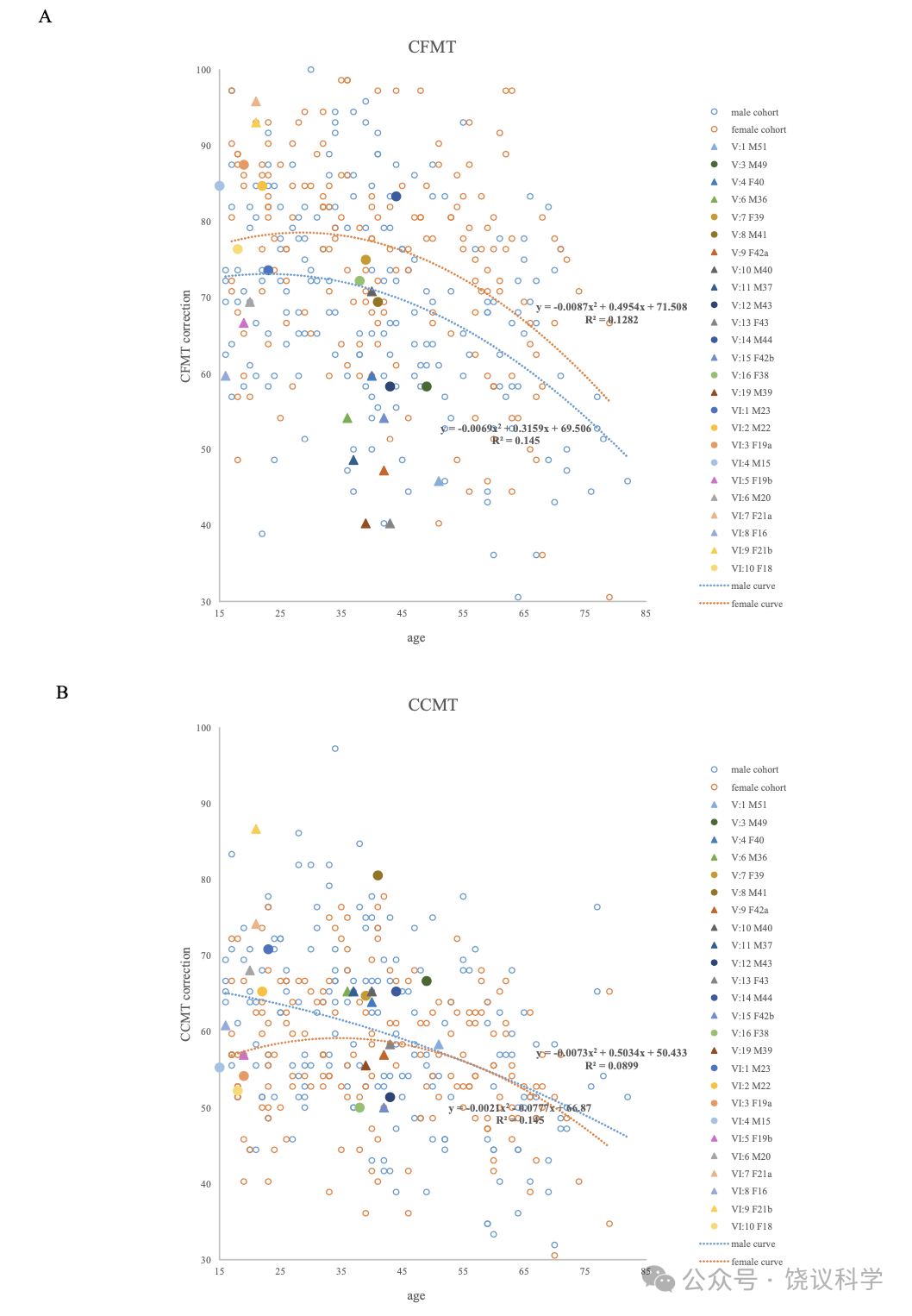
附图S1:Scatterplots of Age against Total Correct for(a)the CFMT-C and(b)the CCMT.
Male control individuals are shown as blue circles; female control individuals are shown as red circles; individuals with daily face recognition problems in Family A are shown as solid triangles; individuals with normal daily face recognition in Family A are shown as solid circles. The curves show the best fitting second-order polynomial relating age to the relevant performance measure with different genders, blue for male, red for female.
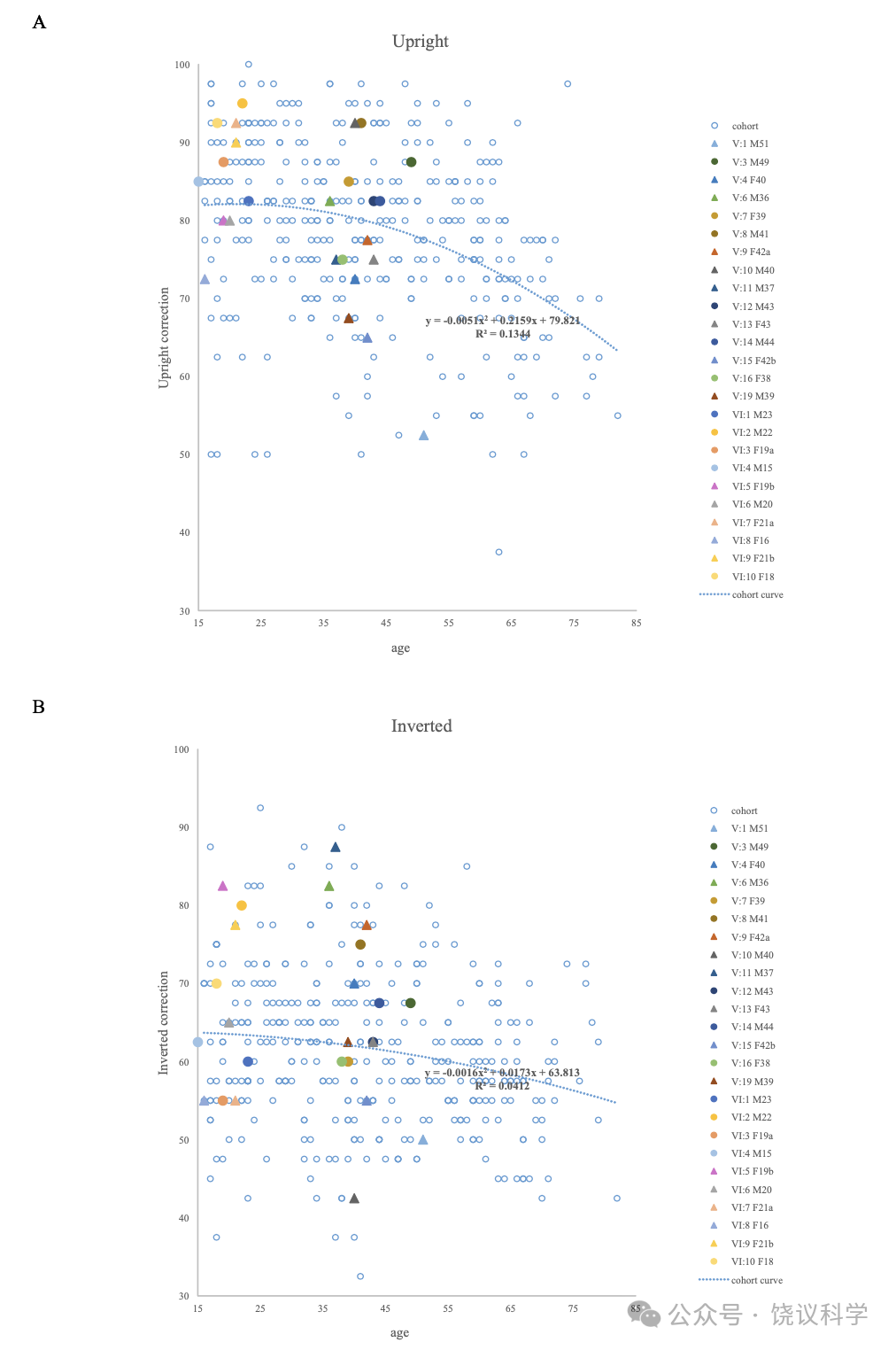
附图S2:Scatterplots of Age against Total Correct for(a)the Upright Face and(b)the Inverted Face.
Control individuals are shown as blue circles; individuals with daily face recognition problems in Family A are shown as solid triangles; individuals with normal daily face recognition in Family A are shown as solid circles. The curves show the best fitting second-order polynomial relating age to the relevant performance measure.
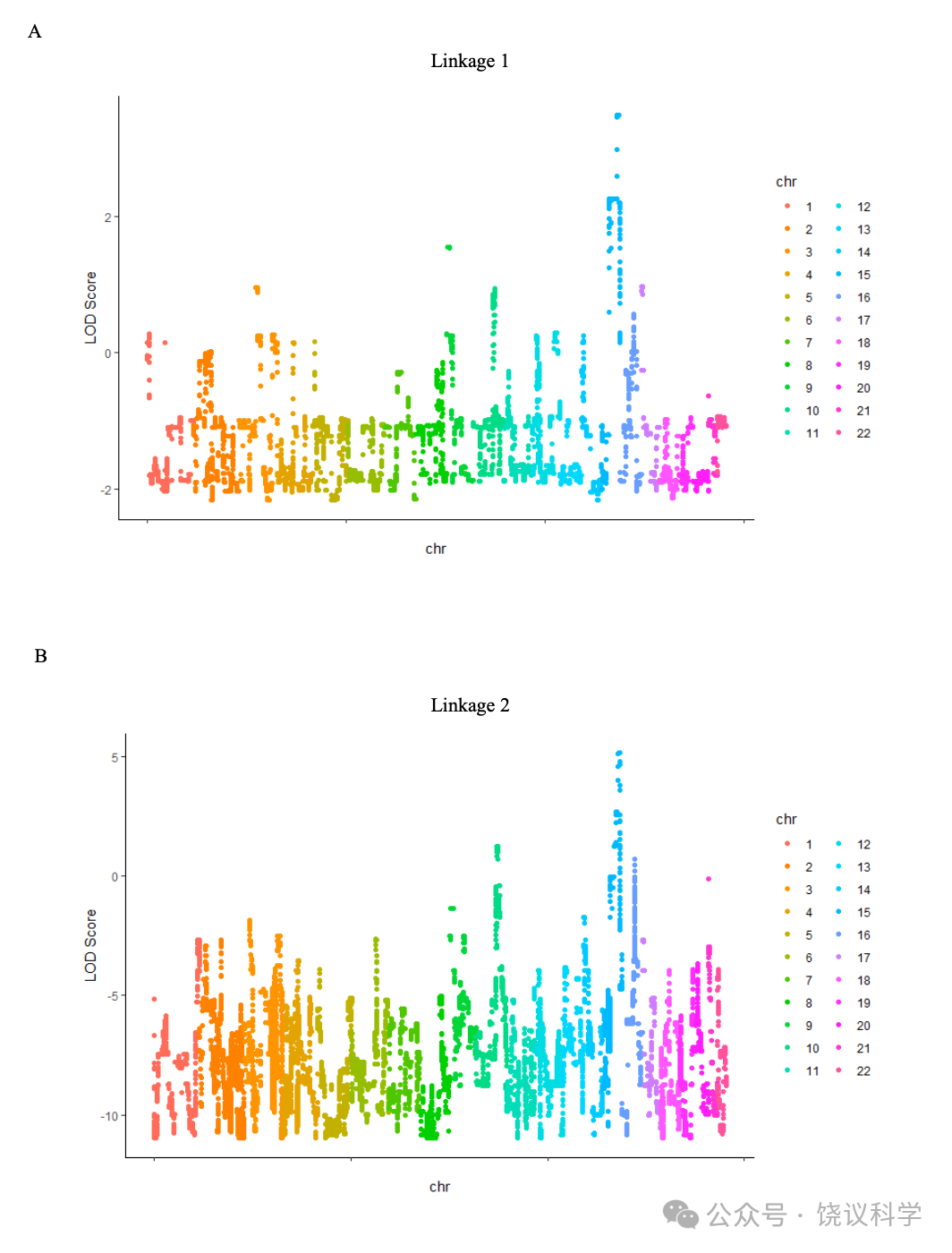
附图S3:Genome-wide Screen Results of CP-susceptibility Loci. Allele-sharing multipoint LOD scores are shown:(a)for linkage 1, and(b)for linkage 2.
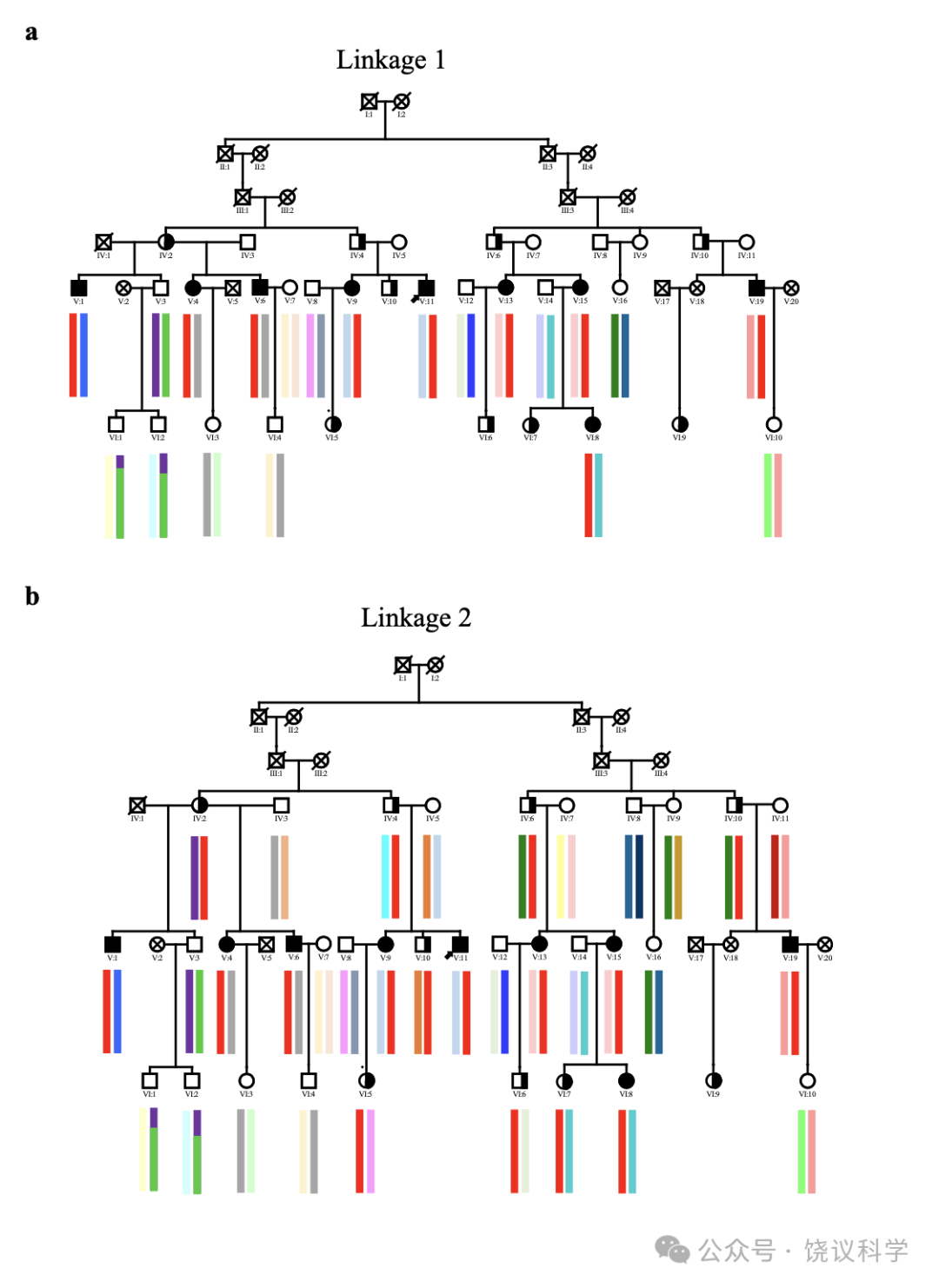
附图S4. Haplotype Analysis for MCR on the Autosomal Dominant CP Locus to 15q26
(a)Genotyping was performed for 20 individuals in linkage 1. Haplotypes for the 3.9 Mb MCR are color-coded, with red indicating co-segregating with CP.
(b)Genotyping was performed for 34 individuals in linkage 2. Haplotypes for the 2.64 Mb MCR are color-coded, with red indicating co-segregating with CP.
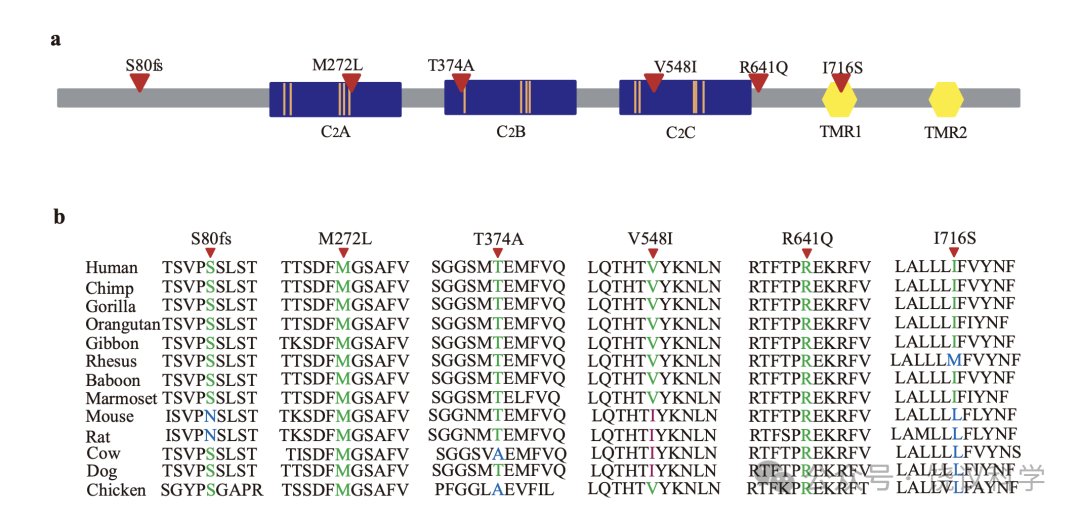
附图S5:The Positions in the MCTP2 Protein Corresponding to MCTP2 Mutations in CPs
(a)The domain plot of the MCTP2 protein and locations of alterations found in CPs. The blue rectangles represent C2 domains, the yellow hexagons represent transmembrane segments; the orange lines indicate putative Ca2+ binding sites; the red triangles indicate amino acid changes.
(b)Evolutionary conservation of amino acid residues in the MCTP2 protein in different species. Amino acids corresponding to mutations found in CP subjects are denoted in different colors. Green:identical to those in the human MCTP2, blue:non-identical to those in human MCTP2, fuchsia:non-identical amino acids but the same as the mutated in human MCTP2. Mutated amino acid residues are indicated by triangles on the top.
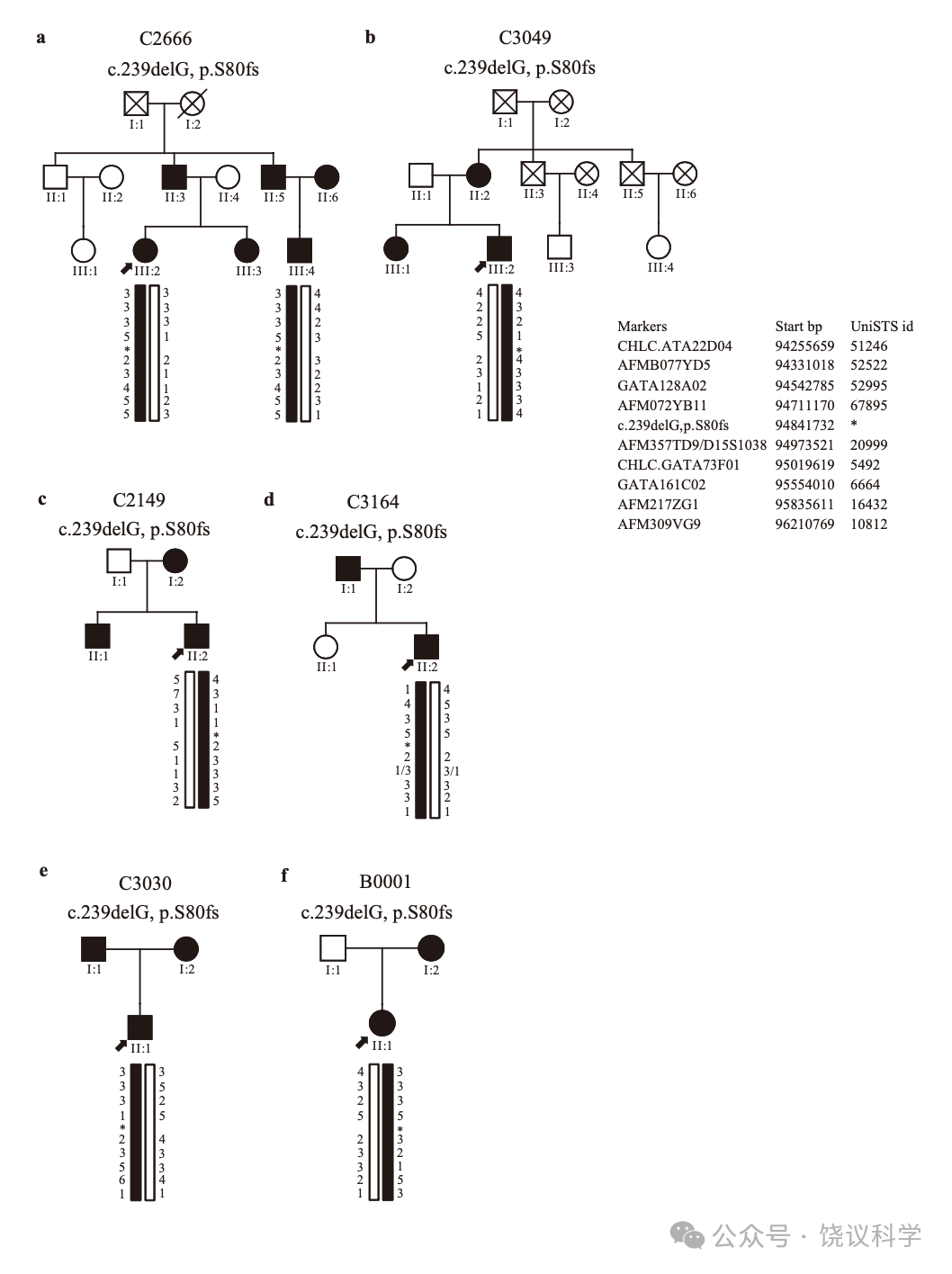
附图S6:Inferred Haplotypes around the c.239delG(p.S80fs)Mutation
These were estimated by comparison to microsatellites data collected on mother-father-offspring trios. Haplotypes coded in black contain the c.239delG(p.S80fs)mutation. The haplotype on the left is paternal and the haplotype on the right is maternal.
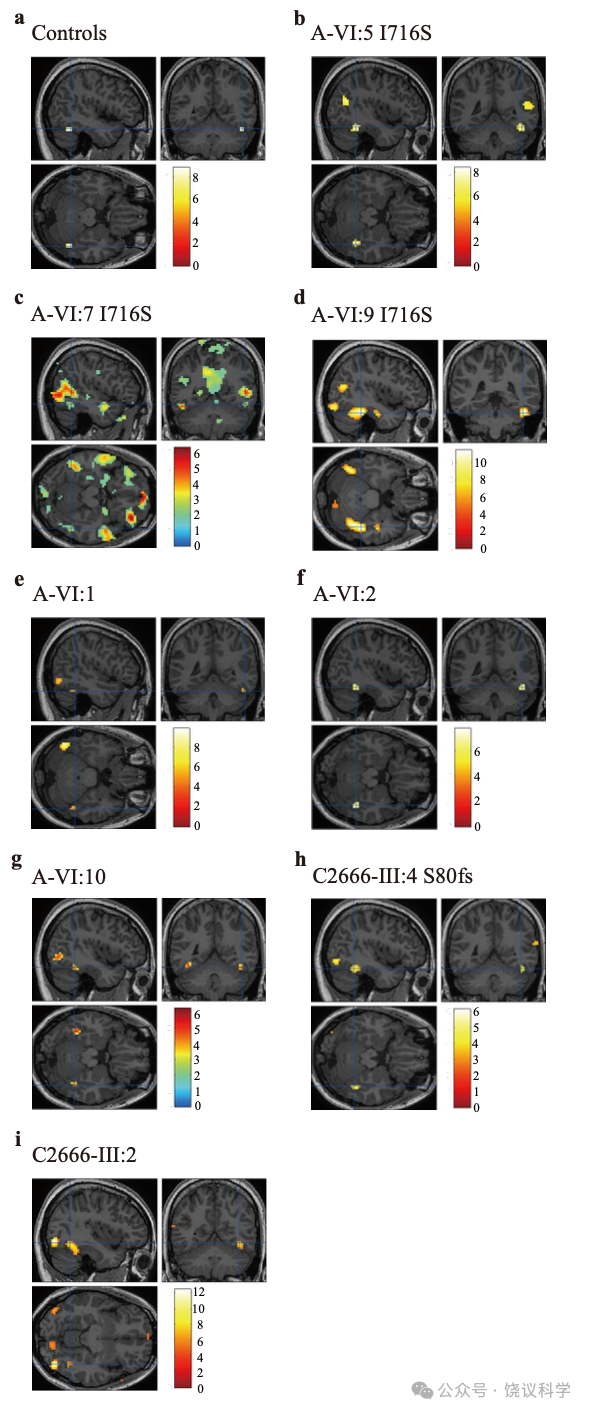
附图S7:Brain fMRI from the Localization Experiment:Identifying the FFA ROI.
Statistical parametric maps by contrasting the response to faces versus non-face stimuli, showing significant face-specific bilateral or unilateral activity in the FFA. Color scale represents statistical values of a one level t test comparing the fMRI signal while subjects viewed blocks of faces versus blocks of non-face stimuli.
(a)rFFA responses in normal controls, pFWE < 0.05.
(b)rFFA responses in A-VI:5 CP member with the I716S mutation, pFWE < 0.05.
(c)rFFA responses in A-VI:7 CP member with the I716S mutation, p < 0.05.
(d)rFFA responses in A-VI:9 CP member with the I716S mutation, pFWE < 0.05.
(e)rFFA responses in A-VI:1 non-CP member, pFWE < 0.05.
(f)rFFA responses in A-VI:2 non-CP member, pFWE < 0.05.
(g)rFFA responses in A-VI:10 non-CP member, p < 0.001.
(h)rFFA responses in C2666-III:4 S80fs CP member, p < 0.0001.
(i)rFFA responses in C2666-III:2 non-CP member, pFWE < 0.05.
表
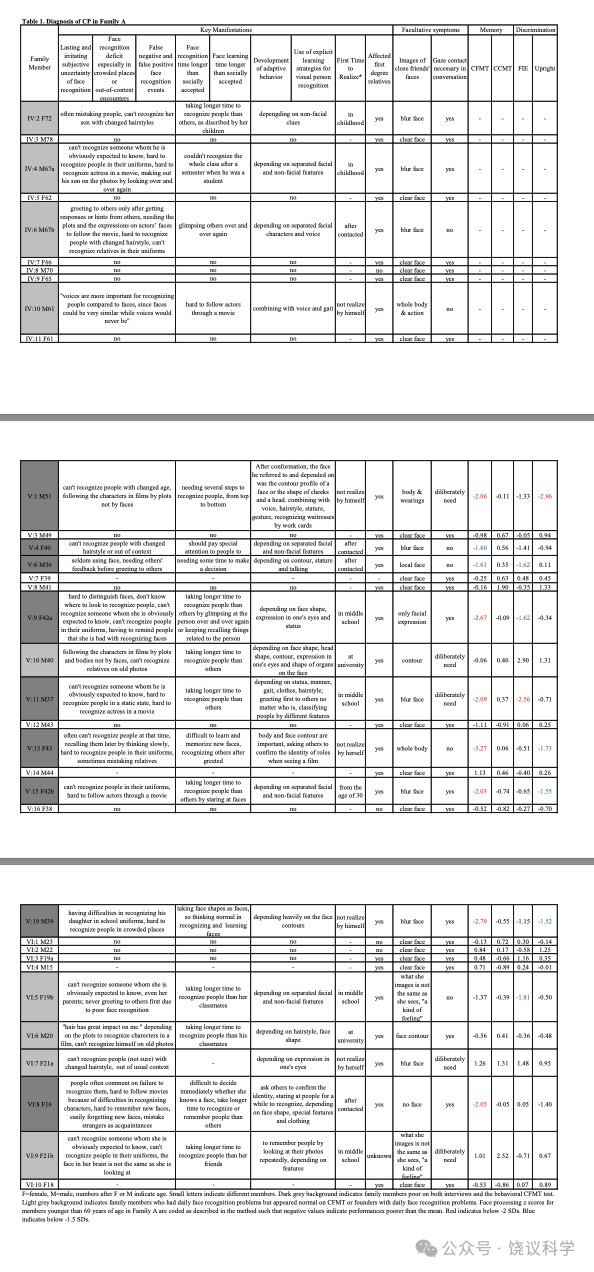
表1. Diagnosis of CP in Family A
F=female, M=male, numbers after F or M indicate age. Small letters indicate different members. Dark grey background indicates family members poor on both interviews and the behavioral CFMT test. Light grey background indicates family members who had daily face recognition problems but appeared normal on CFMT or founders with daily face recognition problems. Face processing Z scores for members younger than 60 years of age in Family A are coded as described in the method such that negative values indicate performances poorer than the mean. Red indicates below -2 SDs. Blue indicates below -1.5 SDs.
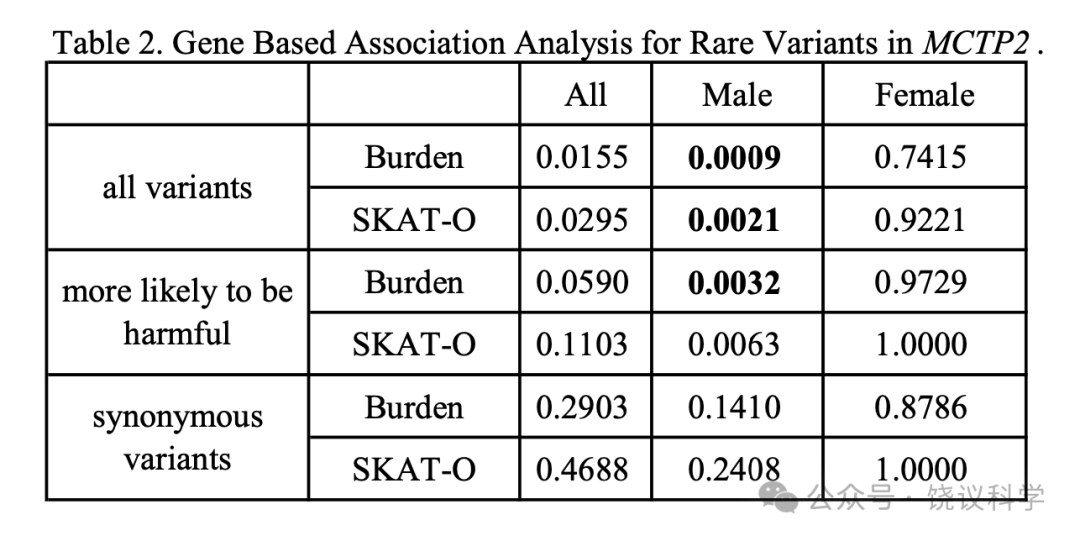
表2. Gene Based Association Analysis for Rare Variants in MCTP2
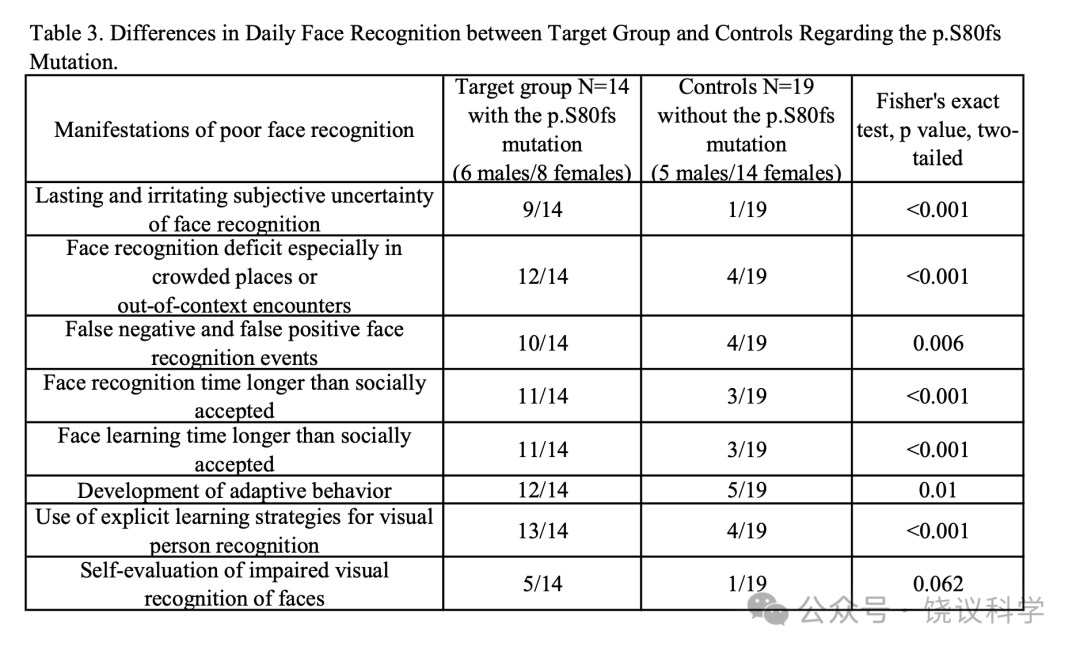
表3. Differences in Daily Face Recognition between Target Group and Controls Regarding the p.S80fs Mutation.
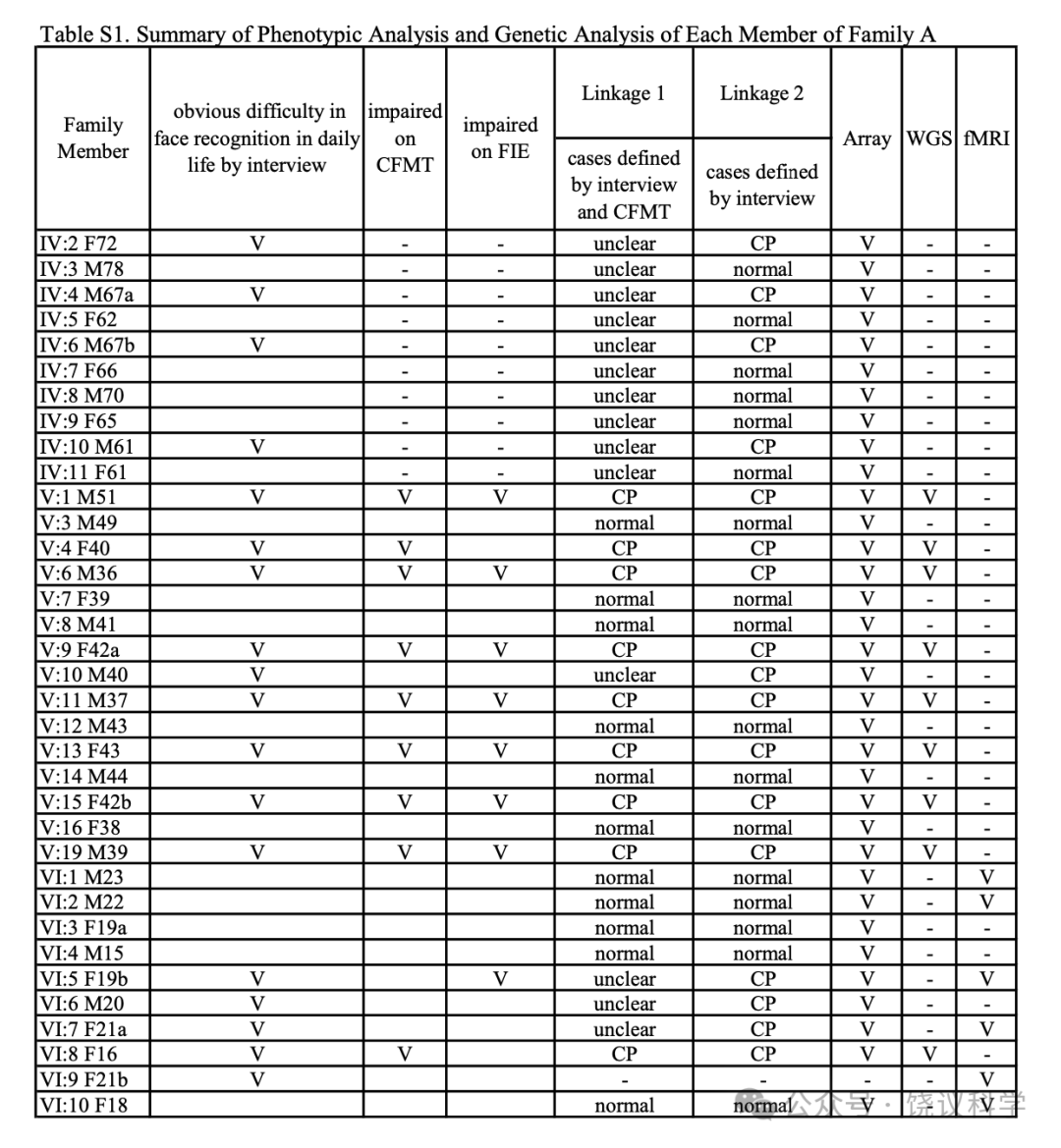
附表S1. Summary of Phenotypic Analysis and Genetic Analysis of Each Member of Family A
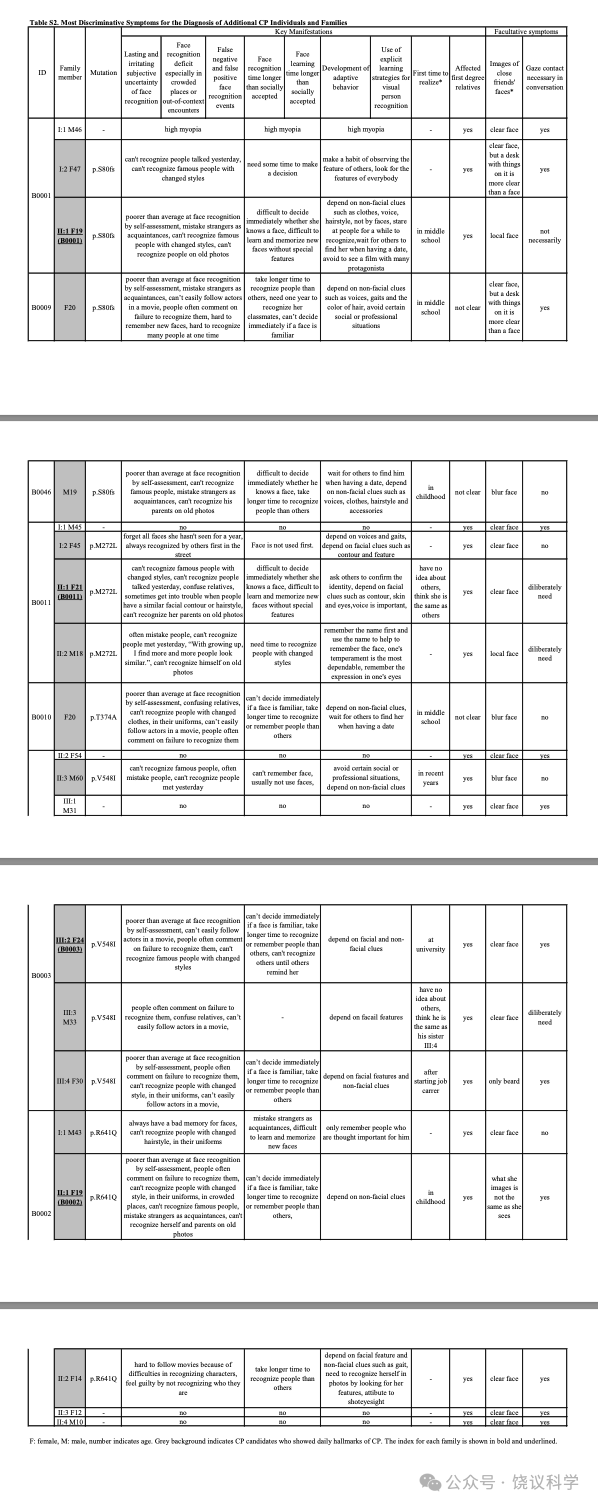
附表S2. Most Discriminative Symptoms for the Diagnosis of Additional CP Individuals and Families
F:female, M:male, number indicates age. Grey background indicates CP candidates who showed daily hallmarks of CP. The index for each family is shown in bold and underlined.
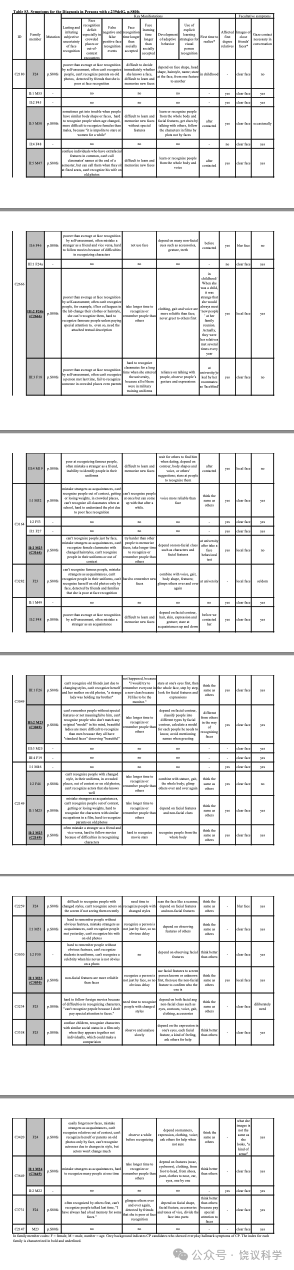
附表S3. Symptoms for the Diagnosis in Persons with c.239delG(p.S80fs)
F:female, M:male, number indicates age. Grey background indicates CP candidates who showed everyday hallmark symptoms of CP. The index for each family is characterized in bold and underlined.
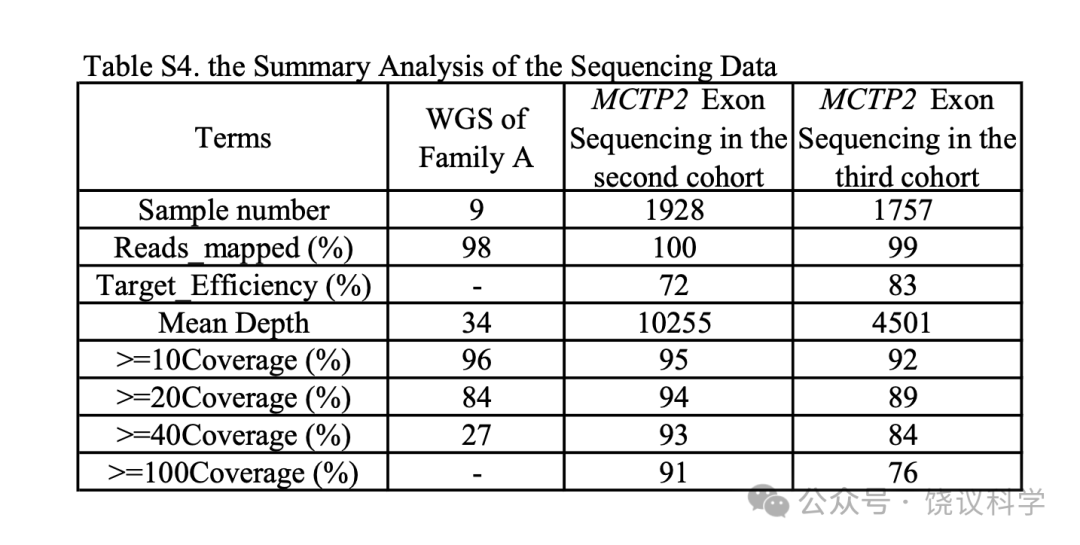
附表S4. the Summary Analysis of the Sequencing Data
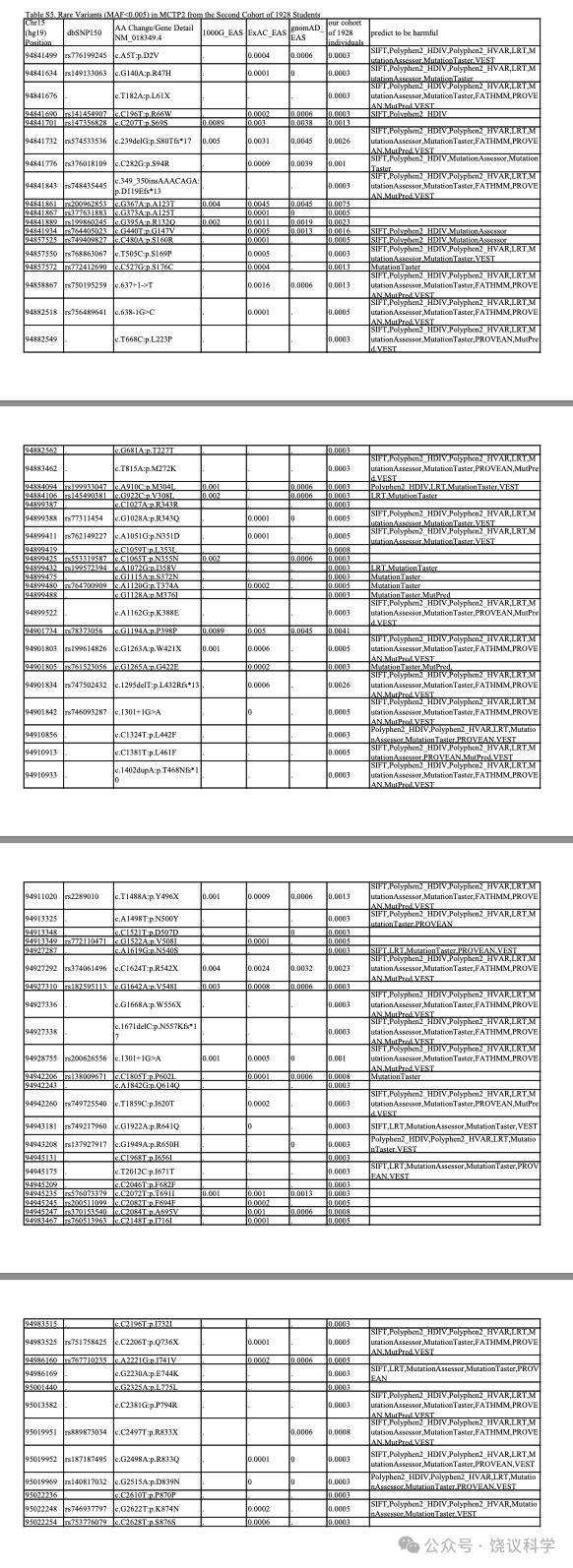
附表S5. Rare Variants(MAF < 0.005)in MCTP2 from a Second Cohort of 1928 Students
* Start Position Based on the MCTP2 RefSeq(hg19, GenBank:NM_018349); # dbSNP build 150
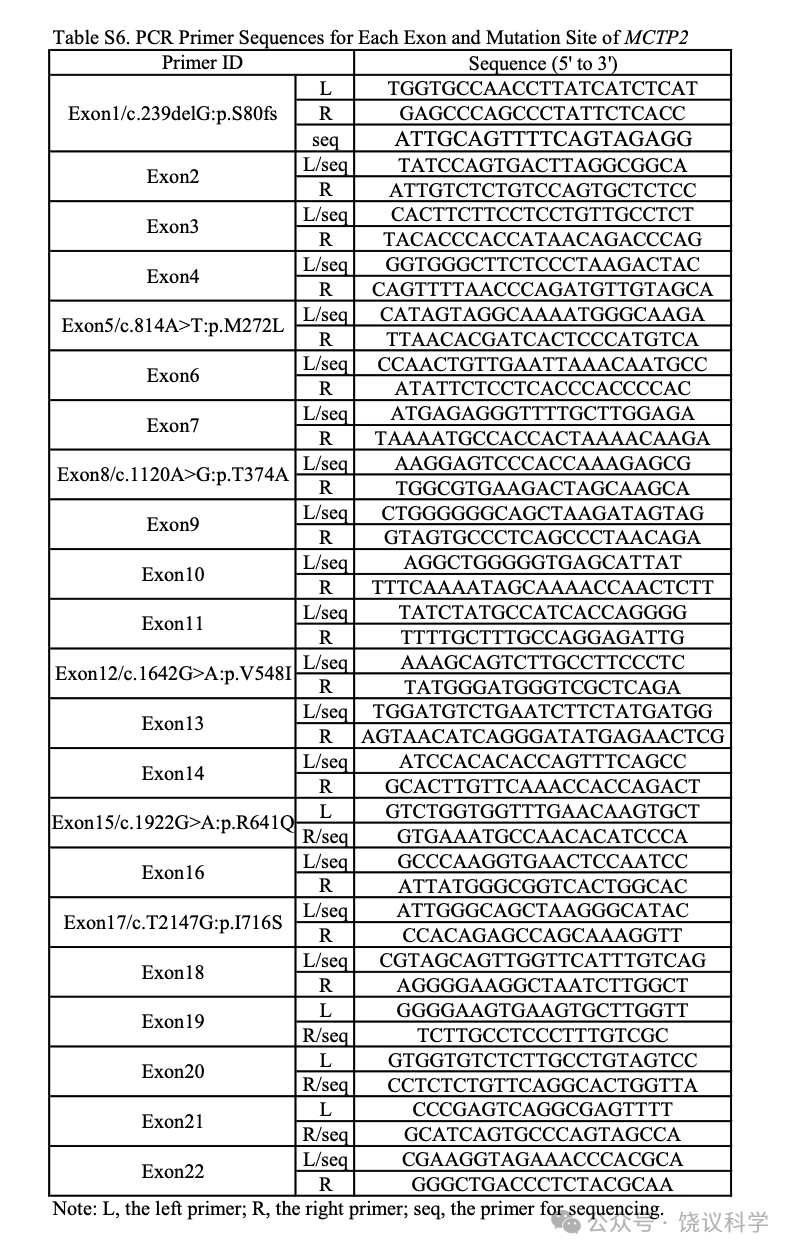
附表S6. PCR Primer Sequences for Each Exon and Mutation Site of MCTP2
L, the left primer; R, the right primer; seq, the primer for sequencing
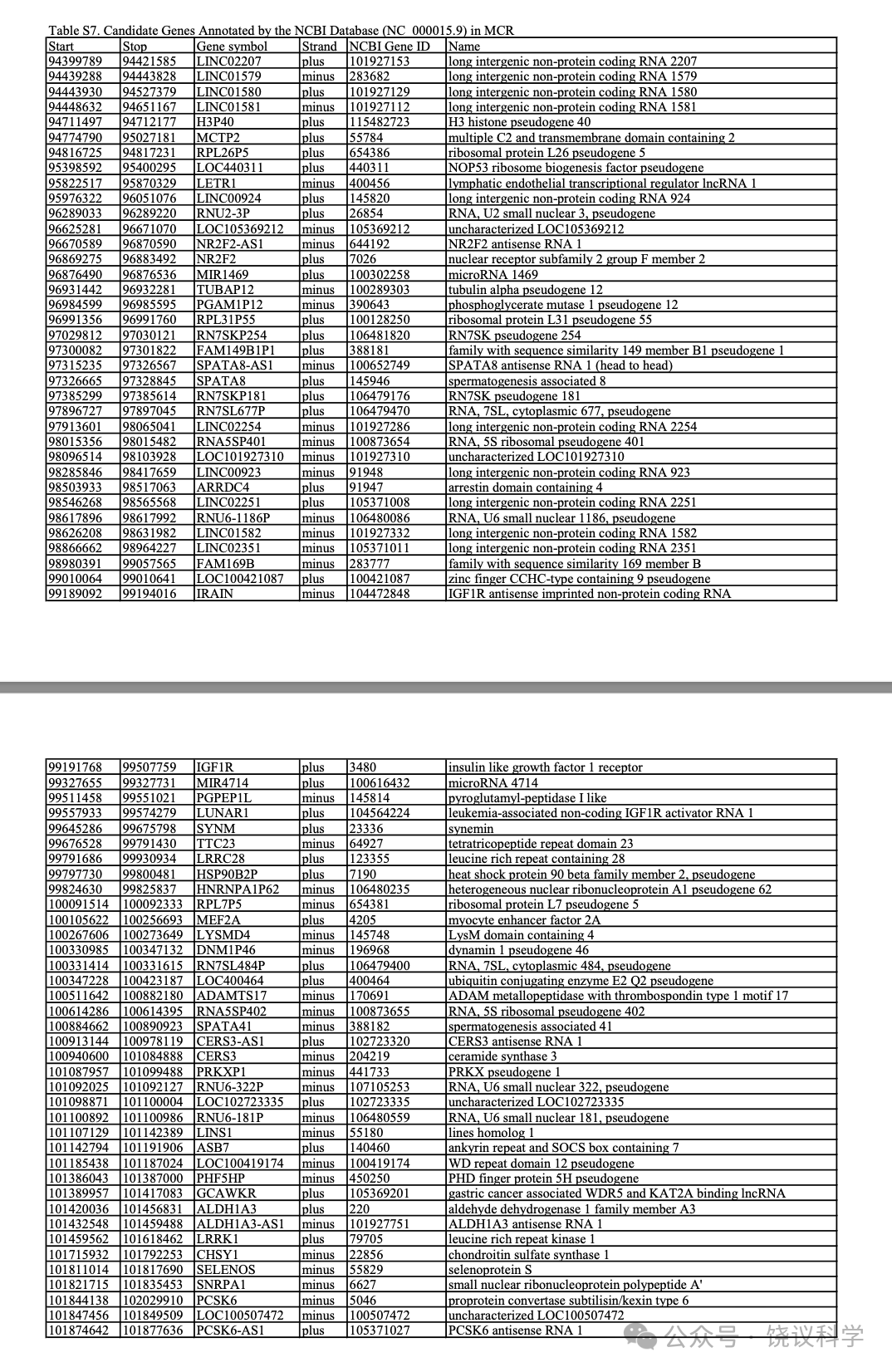
附表S7. Candidate Genes Annotated by the NCBI Database(NC_000015.9)in MCR
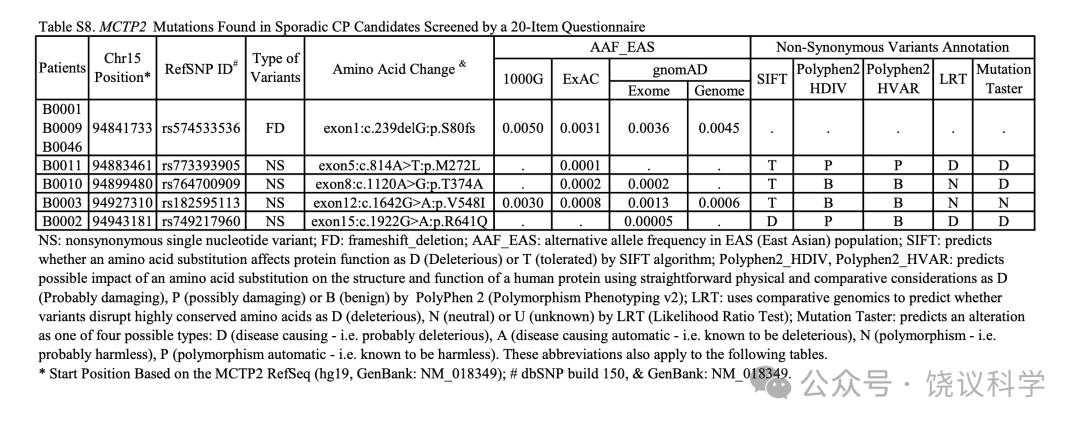
附表S8. MCTP2 Mutations Found in Sporadic CP Candidates Screened by a 20-Item Questionnaire
NS:nonsynonymous single nucleotide variant; FD:frameshift_deletion; AAF_EAS:alternative allele frequency in EAS(East Asian)population; SIFT:predicts whether an amino acid substitution affects protein function as D(Deleterious)or T(tolerated)by SIFT algorithm; Polyphen2_HDIV, Polyphen2_HVAR:predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations as D(Probably damaging), P(possibly damaging)or B(benign)byPolyPhen 2(Polymorphism Phenotyping v2); LRT:uses comparative genomics to predict whether variants disrupt highly conserved amino acids as D(deleterious), N(neutral)or U(unknown)by LRT(Likelihood Ratio Test); Mutation Taster:predicts an alteration as one of four possible types:D(disease causing - i.e. probably deleterious), A(disease causing automatic - i.e. known to be deleterious), N(polymorphism - i.e. probably harmless), P(polymorphism automatic - i.e. known to be harmless). These abbreviations also apply to the following tables. * Start Position Based on the MCTP2 RefSeq(hg19, GenBank:NM_018349); # dbSNP build 150, & GenBank:NM_018349.